Documenti di Didattica
Documenti di Professioni
Documenti di Cultura
A Method For Calibration and Validation Subset Partitioning
Caricato da
Madan LakshmananDescrizione originale:
Titolo originale
Copyright
Formati disponibili
Condividi questo documento
Condividi o incorpora il documento
Hai trovato utile questo documento?
Questo contenuto è inappropriato?
Segnala questo documentoCopyright:
Formati disponibili
A Method For Calibration and Validation Subset Partitioning
Caricato da
Madan LakshmananCopyright:
Formati disponibili
Talanta 67 (2005) 736740
A method for calibration and validation subset partitioning
Roberto Kawakami Harrop Galv o a , M rio C sar Ugulino Araujo b, , Gledson Emdio Jos b , a a e e b , Edvan Cirino Silva b , Teresa Cristina Bezerra Saldanha b Marcio Jos Coelho Pontes e
b a Instituto Tecnol gico de Aeron utica, Divis o de Engenharia Eletr nica, S o Jos dos Campos, S o Paulo, Brazil o a a o a e a Universidade Federal da Paraba, Departamento de Qumica, P.O. Box 5093, Jo o Pessoa, Paraiba 58051-970, Brazil a
Received 18 February 2005; received in revised form 24 March 2005; accepted 24 March 2005 Available online 6 May 2005
Abstract This paper proposes a new method to divide a pool of samples into calibration and validation subsets for multivariate modelling. The proposed method is of value for analytical applications involving complex matrices, in which the composition variability of real samples cannot be easily reproduced by optimized experimental designs. A stepwise procedure is employed to select samples according to their differences in both x (instrumental responses) and y (predicted parameter) spaces. The proposed technique is illustrated in a case study involving the prediction of three quality parameters (specic mass and distillation temperatures at which 10 and 90% of the sample has evaporated) of diesel by NIR spectrometry and PLS modelling. For comparison, PLS models are also constructed by full cross-validation, as well as by using the KennardStone and random sampling methods for calibration and validation subset partitioning. The obtained models are compared in terms of prediction performance by employing an independent set of samples not used for calibration or validation. The results of F-tests at 95% condence level reveal that the proposed technique may be an advantageous alternative to the other three strategies. 2005 Elsevier B.V. All rights reserved.
Keywords: Sample subset partitioning; PLS regression; KennardStone algorithm; NIR spectrometry; Diesel analysis
1. Introduction In multivariate calibration problems involving complex matrices, it can be difcult to reproduce the composition variability of real samples by means of optimized experimental designs [1]. A typical example consists of fuel analysis for the determination of quality parameters such as octane number, cetane index, sulphur content, distillation temperatures, ash point, freezing point, percentage of aromatics and specic mass to name only a few [24]. In such cases, a representative calibration set must be extracted from a pool of real samples. Moreover, validation samples should also be selected to assess the quality of the model and to determine model parameters such as the number of latent variables in PLS regression [5].
Corresponding author. Tel.: +55 83 216 7438; fax: +55 83 216 7437. E-mail address: laqa@quimica.ufpb.br (M.C.U. Araujo).
Several works have addressed the problem of selecting a representative subset from a large pool of samples [69]. In this context, random sampling (RS) is a popular technique because of its simplicity and also because a group of data randomly extracted from a larger set follows the statistical distribution of the entire set. However, RS does not guarantee the representativity of the set, nor does it prevent extrapolation problems [10]. In fact, RS does not ensure that the samples on the boundaries of the set are included in the calibration. An alternative to RS that is often employed is the KennardStone (KS) algorithm. KS is aimed at covering the multidimensional space in a uniform manner by maximizing the Euclidean distances between the instrumental response vectors (x) of the selected samples [912]. In a neural network classication study by Wu et al. [9], KS was found to be superior to RS, as well as to Kohonen self-organizing mapping [13]. The study also showed that KS leads to classication results similar to those obtained by using the more elaborate and time-consuming D-optimal design method [1].
0039-9140/$ see front matter 2005 Elsevier B.V. All rights reserved. doi:10.1016/j.talanta.2005.03.025
R.K.H. Galv o et al. / Talanta 67 (2005) 736740 a
737
It is worth noting that the specic problem of partitioning a pool of real samples into calibration and validation sets for multivariate calibration purposes has not been extensively explored in the literature. Kanduc et al. [10] addressed this problem in a case study involving the prediction of colour properties of a titanium dioxide white pigment from other physical and chemical parameters. The study involved the comparison of RS, KS, Kohonen self-organizing mapping and time-dependent sampling. The models obtained in this manner were compared in terms of their generalization performance in a third prediction set not employed in the modelling procedures. The results revealed that the best predictions were achieved by using KS. However, it should be noticed that the choice of the prediction set was not entirely unbiased in that the prediction samples were extracted from the validation set after the calibration/validation partitioning had already been performed. Moreover, the authors emphasize that an investigation of this problem on a case-by-case basis is always recommended. Despite the comparative advantages of KS over the alternative partitioning methods cited above, a shortcoming of KS in the multivariate calibration context lies in the fact that the statistics of the dependent variable (y) are not taken into account. It could be argued that the inclusion of y-information in the selection process might result in a more effective distribution of calibration samples in the multidimensional space, thus improving the predictive ability and robustness of the resulting model. In the work of Dantas Filho et al. [14], an approach for considering joint xy statistics in the selection of calibration samples was proposed for the purpose of total sulphur determination in diesel samples by NIR spectrometry. However, such an approach was aimed at extracting a reduced subset from the pool of calibration samples, rather than partitioning the available data into calibration and validation. In fact, the analyst was required to provide the calibration and validation sets as a starting point for the sample selection procedure. For this purpose, the calibration/validation partitioning was carried out in a qualitative manner on the basis of a univariate inspection of the reference parameter values followed by an analysis of the residual x and y-variance in the PLS regression. In the present paper, a method for calibration/validation partitioning is proposed to take into account the variability in both x and y dimensions. The method, termed SPXY (Sample set Partitioning based on joint xy distances), extends the KS algorithm by encompassing both x- and y-differences in the calculation of inter-sample distances. For illustration, a multivariate calibration problem involving NIR spectrometric analysis of diesel samples is considered. Three quality parameters are determined, namely specic mass and the distillation temperatures at which 10 and 90% of the sample has evaporated (T10 and T90%). SPXY is compared with KS and RS for the division of modelling data into calibration and validation sets for PLS regression. The performances of the resulting models are compared in terms of root-mean-
square errors calculated in prediction sets not included in the modelling procedures. For the purpose of ensuring the independence of such sets, the prediction samples are randomly extracted from the initial pool of experimental data, before the calibration/validation partitioning procedures. In order to improve the robustness of the error statistics, the study is repeated ve times by resampling the prediction set. The three strategies (PLS-SPXY, PLS-KS, and PLS-RS) are also compared with PLS employing full cross-validation (PLS-CV).
2. Background and theory 2.1. KS algorithm The classic KS algorithm is aimed at selecting a representative subset from a pool of N samples. In order to ensure a uniform distribution of such a subset along the x (instrumental response) data space, KS follows a stepwise procedure in which new selections are taken in regions of the space far from the samples already selected. For this purpose, the algorithm employs the Euclidean distances dx (p, q) between the x-vectors of each pair (p, q) of samples calculated as
J
dx (p, q) =
j=1
[xp (j) xq (j)]2 ;
p, q [1, N]
(1)
For spectral data, xp (j) and xq (j) are the instrumental responses at the jth wavelength for samples p and q, respectively. J denotes the number of wavelengths in the spectra. The selection starts by taking the pair (p1 , p2 ) of samples for which the distance dx (p1 , p2 ) is the largest. At each subsequent iteration, the algorithm selects the sample that exhibits the largest minimum distance with respect to any sample already selected. Such a procedure is repeated until the number of samples specied by the analyst is achieved. 2.2. Proposed SPXY algorithm The proposal of the present paper consists of augmenting the distance dened in Eq. (1) with a distance in the dependent variable (y) space for the parameter under consideration. Such a distance dy (p, q) can be calculated for each pair of samples p and q as dy (p, q) = (yp yq )2 = |yp yq |; p, q [1, N] (2)
In order to assign equal importance to the distribution of the samples in the x and y spaces, distances dx (p, q) and dy (p, q) are divided by their maximum values in the data set. In this manner, a normalized xy distance is calculated as dxy (p, q) = dx (p, q) maxp,q [1,N] dx (p, q) + dy (p, q) ; maxp,q [1,N] dy (p, q) p, q [1, N] (3)
738
R.K.H. Galv o et al. / Talanta 67 (2005) 736740 a
A stepwise selection procedure similar to the KS algorithm can then be applied with dxy (p, q) instead of dx (p, q) alone. The Matlab code for implementation of the proposed SPXY algorithm can be found in Appendix A.
3. Experimental 3.1. Samples The data set consisted of 170 diesel samples that were collected from gas stations in the city of Recife (Pernambuco State, Brazil) and stored in ambar glass asks. 3.2. Reference methods and apparatus The reference values for specic mass and distillation temperatures (T10 and T90%) were obtained according to the ASTM (American Society for Testing and Materials) 4615 and D86 methods, respectively. Specic mass and distillation temperatures were determined by using a Kyoto Electronics DA-130 digital densimeter, and a Herzog HDA 628 automatic distiller, respectively, which were operated according to the recommendations of the manufacturers for optimal working conditions. 3.3. NIR spectra acquisition and pre-processing The spectra were acquired using a FT-NIR/MIR spectrometer Perkin Elmer GX with a spectral resolution of 2 cm1 , 16 scans and an optical path length of 1.0 cm. Only the NIR region in the range 8851600 nm was exploited, because at shorter wavelengths (<885 nm) the signal is too close to the baseline, whereas above 1600 nm the signal saturates the detector. In order to circumvent the problem of systematic variations in the baseline, derivative spectra were calculated with a SavitzkyGolay lter using a 2nd-order polynomial and a 11-point window. Each resulting spectrum had 1431 variables. 3.4. Software Spectrum derivation and PLS modelling were performed with The Unscrambler 7.5 software (CAMO). By using the default settings of the software package, the number of latent variables in the PLS model was determined either by testing on the validation set (PLS-RS, PLS-KS, PLS-SPXY) or by full cross-validation (PLS-CV). RS, KS, and SPXY routines were implemented in Matlab 6.1. The division of the 170 samples into calibration, validation, and prediction sets was carried out in the following manner. Initially, 50 prediction samples were extracted from the full set in a random manner to simulate the analysis of a batch of real unknown samples. The remaining 120 samples were divided into calibration and validation sets of 70 and 50 elements, respectively, by using
the three selection methods to be compared (RS, KS, and SPXY). In order to improve the statistical signicance of the comparison, the extraction of the prediction set and the subsequent partitioning of the remaining samples into calibration and validation by RS, KS, and SPXY was repeated ve times. In this manner, the four modelling strategies (PLS-RS, PLSKS, PLS-SPXY, and PLS-CV) were tested with ve different prediction sets. Because of the random nature of the RS method, special care was taken to improve the statistical signicance of the PLS-RS results. For this purpose, ve RS calibration/validation partitions were performed for each of the ve extractions of the prediction set. In this manner, 5 5 = 25 evaluations of PLS-RS were carried out. For each diesel quality parameter, the predictive ability of PLS-RS, PLS-KS, PLS-SPXY, and PLS-CV were compared in terms of an overall root-mean-square error of prediction (RMSEP). Such an RMSEP statistic was dened for PLS-KS, PLS-SPXY, and PLS-CV as RMSEP = 1 I M
I M
(yi,m yi,m )2
i=1 m=1
(4)
where yi,m and yi,m are the reference and predicted values of the parameter under consideration in the mth prediction sample (m = 1, . . ., M) of the ith prediction set (i = 1, . . ., I). As explained above, M = 50, and I = 5 were employed in this work. The RMSEP calculation for PLS-RS also embodied the RS repetitions as RMSEP = 1 I KM
I K M
(yi,k,m yi,k,m )2
i=1 k=1 m=1
(5)
where index k = 1, . . ., K refers to each of the ve (K = 5) calibration/validation divisions by RS. The statistical signicance of differences between RMSEP values were assessed by using an F-test for a condence level of 95%. It is worth noting that the RMSEP calculation for PLS-RS involves ve times more degrees of freedom than the respective calculation for each other modelling strategy (PLS-KS, PLS-SPXY, and PLS-CV).
4. Results and discussion The original spectra of the 170 diesel samples analyzed by NIR spectrometry are presented in Fig. 1a. Such spectra display baseline features that were corrected by derivation with a SavitzkyGolay lter. Fig. 1b shows the resulting derivative spectra, which were employed throughout the work. Table 1 presents the RMSEP results of the four modelling strategies for each parameter under study. As regards the comparison of PLS-KS, PLS-RS, and PLSCV performances, it can be seen that PLS-CV yielded the smallest RMSEP for specic mass, whereas PLS-RS yielded
R.K.H. Galv o et al. / Talanta 67 (2005) 736740 a Table 1 RMSEP results obtained for each modelling strategy Parameter Specic mass T10% (186.6269.9 C) T90% (317.2385.5 C) (830864 kg m3 ) PLS-KS 1.8 5.5 4.7 PLS-RS 1.8 5.4 4.4 PLS-CV 1.6 5.5 4.5
739
PLS-SPXY 1.7 5.3 4.0
The range of each parameter in the data set is indicated in parenthesis.
culations, is considerably more demanding in computational terms. SPXY was successfully employed with PLS regression for NIR spectrometric determination of specic mass and T10, and T90% distillation temperatures in diesel samples. The results showed that the proposed method may be an advantageous alternative to divide modelling data into calibration and validation sets for PLS regression in comparison with KS, RS, or full cross-validation.
Acknowledgments The authors thank FINEP-CTPETRO (Grant 0652/00) science funding program, PROCAD/CAPES (Grant 0064/01-7), PRONEX/CNPq (Grant 015/98), and FAPESP (Grant 03/09433-5) for partial nancial support. The research fellowships granted by the Brazilian agency CNPq are also gratefully acknowledged. The authors are also indebted to the Laborat rio de Combustveis of the Departamento o de Engenharia Qumica of the Universidade Federal de Pernambuco for providing the diesel samples and reference values for this work.
Fig. 1. Original (a) and derivative (b) NIR spectra of the 170 diesel samples.
the smallest RMSEP for T10 and T90%. However, the F-test reveals that the only signicant differences favour PLS-CV over the other two strategies for specic mass. On the other hand, the RMSEP of the proposed technique (PLS-SPXY) is smaller than the corresponding values of the other strategies in almost all cases. The only exception consists of PLS-CV for specic mass, but even in this case the difference with respect to PLS-SPXY is not signicant at the adopted condence level (95%) of the F-test. In fact, PLSSPXY is favoured over the other strategies in all signicant F-test comparisons (against PLS-KS, PLS-RS, and PLS-CV in T90%).
Appendix A. Matlab implementation of the proposed SPXY algorithm In this Matlab function, X and y are the instrumental response matrix (independent variables) and the column vector of parameter values (dependent variable), respectively. Ncal is the number of objects to be selected for the calibration set. The indexes of the selected objects are returned in vector m.
function m = spxy(X,y,Ncal) dminmax = zeros(1,Ncal); % Inicializes the vector of minimum distances. M = size(X,1); % Number of objects samples = 1:M; Dx = zeros(M,M); % Inicializes the matrix of X-distances. Dy = zeros(M,M); % Inicializes the matriz de y-distances. for i = 1:M-1 xa = X(i,:); ya = y(i,:); for j = i + 1:M xb = X(j,:); yb = y(j,:); Dx(i, j) = norm(xaxb); Dy(i, j) = norm(yayb); end end
5. Conclusions This paper proposed a method to divide modelling data into calibration and validation sets for multivariate calibration. The method, termed SPXY, employs a partitioning algorithm that takes into account the variability in both xand y-spaces. In this manner, the multidimensional space may be covered more effectively in comparison with partitioning schemes based on x-information alone (such as the KennardStone (KS) algorithm) or random sampling (RS). As a result, improvements on the prediction performance of the resulting PLS models may be attained. In terms of computational workload, SPXY is comparable with KS, in the sense that both algorithms employ simple distance calculations. In contrast, full cross-validation, which is often used in PLS cal-
740
R.K.H. Galv o et al. / Talanta 67 (2005) 736740 a
Dxmax = max(max(Dx)); Dymax = max(max(Dy)); D = Dx/Dxmax + Dy/Dymax; % Combines the X and y distances. % D is an upper triangular matrix. % D(i,j) is the distance between objects i and j (j > i). [maxD,index row] = max(D); % maxD is a row vector containing the largest element for each column of D. % index row is the row in which the largest element of the column if found. [dummy,index column] = max(maxD); % index column is the column containing the largest element of matrix D. m(1) = index row(index column); m(2) = index column; for i = 3:Ncal pool = setdiff(samples,m); % Pool is the index set of the samples that have not been selected yet. dmin = zeros(1,M i + 1); % dmin will store the minimum distance of each sample in pool with respect to the previously selected samples. for j = 1:(M i + 1) indexa = pool(j); d = zeros(1,i-1); for k = 1:(i 1) indexb = m(k); if indexa < indexb d(k) = D(indexa,indexb); else d(k) = D(indexb,indexa); end end dmin(j) = min(d); end % At each iteration, the sample with the largest dmin value is selected. [dummy,index] = max(dmin); m(i) = pool(index); end
References
[1] D.L. Massart, B.G.M. Vandeginste, L.M.C. Buydens, S. De Jong, P.J. Lewi, J. Smeyers-Verbeke, Handbook of Chemometrics and Qualimetrics: Part A, Elsevier, Amsterdam, 1997. [2] C.T. Manseld, B.N. Barman, Anal. Chem. 71 (1999) 81R. [3] M.D. Judge, Talanta 62 (2004) 675. [4] M.P. Gomez-Carracedo, J.M. Andrade, M.A. Calvino, D. Prada, E. Fernandez, S. Muniategui, Talanta 60 (2003) 1051. [5] K.R. Beebe, R.J. Pell, B. Seasholtz, ChemometricsA Pratical Guide, Wiley, New York, 1998. [6] M. Daszykowski, B. Walczak, D.L. Massart, Anal. Chim. Acta 468 (2002) 91. [7] Y. Tominaga, Chemometr. Intell. Lab. Syst. 43 (1998) 157. [8] F. Sales, A. Rius, M.P. Callao, F.X. Rius, Talanta 52 (2000) 329. [9] W. Wu, B. Walczak, D.L. Massart, S. Heuerding, F. Erni, I.R. Last, K.A. Prebble, Chemometr. Intell. Lab. Syst. 33 (1996) 35. [10] K.R. Kanduc, J. Zupan, N. Majcen, Chemometr. Intell. Lab. Syst. 65 (2003) 221. [11] R.W. Kennard, L.A. Stone, Technometrics 11 (1969) 137. [12] E. Bouveresse, C. Hartmann, D.L. Massart, I.R. Last, K.A. Prebble, Anal. Chem. 68 (1996) 982. [13] L.F. Capitan-Vallvey, N. Navas, M. del Olmo, V. Consonni, R. Todeschini, Talanta 52 (2000) 1069. [14] H.A.D. Dantas Filho, R.K.H. Galv o, M.C.U. Ara jo, E.C. a u Silva, T.C.B. Saldanha, G.E. Jos , C. Pasquini, I.M. Raimundo e Jr., J.J.R. Rohwedder, Chemometr. Intell. Lab. Syst. 72 (2004) 83.
Potrebbero piacerti anche
- Chemometrics and Intelligent Laboratory Systems: 10.1016/j.chemolab.2016.07.011Documento26 pagineChemometrics and Intelligent Laboratory Systems: 10.1016/j.chemolab.2016.07.011JessicaNessuna valutazione finora
- Averaged and Weighted Average Partial Least Squares: M.H. Zhang, Q.S. Xu, D.L. MassartDocumento11 pagineAveraged and Weighted Average Partial Least Squares: M.H. Zhang, Q.S. Xu, D.L. MassartAnonymous cVJKeIOLBNNessuna valutazione finora
- Urkedal,' A.T.: PaperDocumento16 pagineUrkedal,' A.T.: PaperRosa K Chang HNessuna valutazione finora
- Fitting Nonlinear Calibration Curves No Models PerDocumento17 pagineFitting Nonlinear Calibration Curves No Models PerLuis LahuertaNessuna valutazione finora
- Va41 3 405Documento6 pagineVa41 3 405bdalcin5512Nessuna valutazione finora
- Validation of The Calibration Procedure in Atomic Absorption Spectrometric MethodsDocumento10 pagineValidation of The Calibration Procedure in Atomic Absorption Spectrometric MethodsaureaborosNessuna valutazione finora
- Bootstrap confidence intervals for reservoir model selection techniquesDocumento25 pagineBootstrap confidence intervals for reservoir model selection techniquesregarcia81Nessuna valutazione finora
- Ranova Help Tcm18-242064Documento20 pagineRanova Help Tcm18-242064BruzziNessuna valutazione finora
- Prediction Methods in The Presence of MulticollinearityDocumento17 paginePrediction Methods in The Presence of MulticollinearityKhaja Riazuddin Nawaz MohammedNessuna valutazione finora
- Results and Conclusions: 9.1 Summary of The ResultsDocumento8 pagineResults and Conclusions: 9.1 Summary of The ResultsNova MejisienNessuna valutazione finora
- Using Additional Standards For Increasing The Accuracy of Quantitative Chromatographic AnalysisDocumento10 pagineUsing Additional Standards For Increasing The Accuracy of Quantitative Chromatographic AnalysisTiar_Rahman_9553Nessuna valutazione finora
- A Comparison of SIFT, PCA-SIFT and SURF: Computer Graphics Lab, Chonbuk National University, Jeonju 561-756, South KoreaDocumento10 pagineA Comparison of SIFT, PCA-SIFT and SURF: Computer Graphics Lab, Chonbuk National University, Jeonju 561-756, South Koreamatkhau01Nessuna valutazione finora
- Performance Indication Improvement For A Proficiency TestingDocumento5 paginePerformance Indication Improvement For A Proficiency TestingManvel EnriqueNessuna valutazione finora
- Estimating Replicability of Classifier Learning ExperimentsDocumento11 pagineEstimating Replicability of Classifier Learning Experimentssmunoz443x9066Nessuna valutazione finora
- Ball in 1992Documento23 pagineBall in 1992Hossein HaddadpourNessuna valutazione finora
- Cross ValidationDocumento6 pagineCross ValidationSantosh ButleNessuna valutazione finora
- Machine Learning Book Review: An Algorithmic PerspectiveDocumento4 pagineMachine Learning Book Review: An Algorithmic PerspectivePrajwal GowdaNessuna valutazione finora
- SVM Quantile DNADocumento9 pagineSVM Quantile DNAXiao YaoNessuna valutazione finora
- Cross Validation LN 12Documento11 pagineCross Validation LN 12M S PrasadNessuna valutazione finora
- Cross Validation LN 12Documento11 pagineCross Validation LN 12M S PrasadNessuna valutazione finora
- BAYREUTHDocumento15 pagineBAYREUTHAjitNessuna valutazione finora
- Algorithms-09-00046 (Path Based Four Uci Datasets)Documento13 pagineAlgorithms-09-00046 (Path Based Four Uci Datasets)Khin MyintNessuna valutazione finora
- Niels-Peter Vest Nielsen, 1998Documento19 pagineNiels-Peter Vest Nielsen, 1998Laura MonicaNessuna valutazione finora
- An Efficient Method For Paired-Comparison: D. Amnon Silverstein Joyce E. FarrellDocumento15 pagineAn Efficient Method For Paired-Comparison: D. Amnon Silverstein Joyce E. FarrellSharif UllahNessuna valutazione finora
- CHEMOMETRICS and STATISTICS Multivariate Classification Techniques-21-27Documento7 pagineCHEMOMETRICS and STATISTICS Multivariate Classification Techniques-21-27Jose GarciaNessuna valutazione finora
- K Means SpectralDocumento10 pagineK Means SpectralEthan Carres HidalgoNessuna valutazione finora
- 1 s2.0 S0003267017306967 MainDocumento11 pagine1 s2.0 S0003267017306967 Mainmuhammad reza mahendraNessuna valutazione finora
- F.-M. Schleif, T. Villmann and B. Hammer - Prototype Based Fuzzy Classification in Clinical ProteomicsDocumento20 pagineF.-M. Schleif, T. Villmann and B. Hammer - Prototype Based Fuzzy Classification in Clinical ProteomicsGrettszNessuna valutazione finora
- Zhong - 2005 - Efficient Online Spherical K-Means ClusteringDocumento6 pagineZhong - 2005 - Efficient Online Spherical K-Means ClusteringswarmbeesNessuna valutazione finora
- Dechter Constraint ProcessingDocumento40 pagineDechter Constraint Processingnani subrNessuna valutazione finora
- Remotesensing 09 01113 v2Documento24 pagineRemotesensing 09 01113 v2Débora PlácidoNessuna valutazione finora
- Comparative Studies On Some Metrics For External Validation of QSPR ModelsDocumento13 pagineComparative Studies On Some Metrics For External Validation of QSPR ModelsPace RaditNessuna valutazione finora
- Gaussian Kernel Optimization For Pattern Classification: Jie Wang, Haiping Lu, Juwei LuDocumento28 pagineGaussian Kernel Optimization For Pattern Classification: Jie Wang, Haiping Lu, Juwei LuAnshul SuneriNessuna valutazione finora
- A Hybrid Algorithm Based On Cuckoo Search and Differential Evolution For Numerical OptimizationDocumento8 pagineA Hybrid Algorithm Based On Cuckoo Search and Differential Evolution For Numerical OptimizationrasminojNessuna valutazione finora
- A Distance-Based Kernel For Classification Via Support Vector Machines - PMC-19Documento1 paginaA Distance-Based Kernel For Classification Via Support Vector Machines - PMC-19jefferyleclercNessuna valutazione finora
- OPTIMIZE PARAMETER ESTIMATION IN GEOTECHNICAL MODELINGDocumento23 pagineOPTIMIZE PARAMETER ESTIMATION IN GEOTECHNICAL MODELINGHUGINessuna valutazione finora
- Sparkscore: Leveraging Apache Spark For Distributed Genomic InferenceDocumento8 pagineSparkscore: Leveraging Apache Spark For Distributed Genomic InferenceMónicaPitaLópezNessuna valutazione finora
- Journal of Petroleum Science and Engineering: Kyungbook Lee, Jungtek Lim, Jonggeun Choe, Hyun Suk LeeDocumento11 pagineJournal of Petroleum Science and Engineering: Kyungbook Lee, Jungtek Lim, Jonggeun Choe, Hyun Suk LeeYarisol Rivera ChàvezNessuna valutazione finora
- Alehandro Lumentah 210211010188 Assignment09Documento10 pagineAlehandro Lumentah 210211010188 Assignment09Alex FredNessuna valutazione finora
- Prediction Under Uncertainty in Reservoir Modeling: Mike Christie, Sam Subbey, Malcolm SambridgeDocumento8 paginePrediction Under Uncertainty in Reservoir Modeling: Mike Christie, Sam Subbey, Malcolm SambridgeDannyNessuna valutazione finora
- Model Reduction High DimensionDocumento20 pagineModel Reduction High DimensionLei TuNessuna valutazione finora
- Genetic Algorithm-Based Clustering TechniqueDocumento11 pagineGenetic Algorithm-Based Clustering TechniqueArmansyah BarusNessuna valutazione finora
- Final Project ReportDocumento18 pagineFinal Project ReportjstpallavNessuna valutazione finora
- Gaussian Process Landmarking On Manifolds: Tingran Gao, Shahar Z. Kovalsky, Ingrid DaubechiesDocumento30 pagineGaussian Process Landmarking On Manifolds: Tingran Gao, Shahar Z. Kovalsky, Ingrid DaubechiesmeoxauNessuna valutazione finora
- Algorithms 10 00105Documento14 pagineAlgorithms 10 00105kamutmazNessuna valutazione finora
- Analysis&Comparisonof Efficient TechniquesofDocumento5 pagineAnalysis&Comparisonof Efficient TechniquesofasthaNessuna valutazione finora
- Estimates of Uncertainty of Measurement From Proficiency Testing Data: A Case StudyDocumento7 pagineEstimates of Uncertainty of Measurement From Proficiency Testing Data: A Case StudyChuckPeter MartinorrisNessuna valutazione finora
- Improving Expression Data Mining Through ClusterDocumento4 pagineImproving Expression Data Mining Through ClusterYuhefizar Ephi LintauNessuna valutazione finora
- Ecient Leave-One-Out Cross-Validation of Kernel Fisher Discriminant Classi'ersDocumento8 pagineEcient Leave-One-Out Cross-Validation of Kernel Fisher Discriminant Classi'ersMohammad TaheriNessuna valutazione finora
- Analytical Chemistry. - Curvas de Calibracion, Bondad de Ajuste, PonderaciónDocumento8 pagineAnalytical Chemistry. - Curvas de Calibracion, Bondad de Ajuste, Ponderaciónhugoboss_darkNessuna valutazione finora
- Number of Analytes in A PanelDocumento3 pagineNumber of Analytes in A Panelshakthi jayanthNessuna valutazione finora
- Active Learning of Ternary Alloy Structures and EnergiesDocumento22 pagineActive Learning of Ternary Alloy Structures and Energiesbmalki68Nessuna valutazione finora
- (VK) 25. Bayesian Node LocalisationDocumento4 pagine(VK) 25. Bayesian Node LocalisationHarshNessuna valutazione finora
- Implementation of Bayesian Tests Pbayes and Dbayes For Randomized Block Design in R Code.Documento7 pagineImplementation of Bayesian Tests Pbayes and Dbayes For Randomized Block Design in R Code.IJAERS JOURNALNessuna valutazione finora
- Ijaiem 2014 05 20 056Documento11 pagineIjaiem 2014 05 20 056International Journal of Application or Innovation in Engineering & ManagementNessuna valutazione finora
- H-P Discretisation in Three DimensionsDocumento13 pagineH-P Discretisation in Three DimensionsMario Galindo QueraltNessuna valutazione finora
- Bickel and Levina 2004Documento28 pagineBickel and Levina 2004dfaini12Nessuna valutazione finora
- Journal of Chromatography A: David Pfister, Fabian Steinebach, Massimo MorbidelliDocumento9 pagineJournal of Chromatography A: David Pfister, Fabian Steinebach, Massimo MorbidelliJames EdwardsNessuna valutazione finora
- 2001, Pena, PrietoDocumento25 pagine2001, Pena, Prietomatchman6Nessuna valutazione finora
- Seed Enterprises January 2011Documento94 pagineSeed Enterprises January 2011Madan LakshmananNessuna valutazione finora
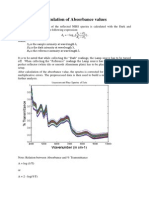
- Calculation of Absorbance ValuesDocumento1 paginaCalculation of Absorbance ValuesMadan LakshmananNessuna valutazione finora
- Which data points go into the training set for molecular modelingDocumento4 pagineWhich data points go into the training set for molecular modelingMadan LakshmananNessuna valutazione finora
- Smoothing and Differentiation of Data by Simplified Least Squares ProceduresDocumento13 pagineSmoothing and Differentiation of Data by Simplified Least Squares ProceduresMadan LakshmananNessuna valutazione finora
- Selecting Arrester MCOV-UcDocumento9 pagineSelecting Arrester MCOV-Ucjose GonzalezNessuna valutazione finora
- MF1547Front Linkage - Seat PDFDocumento18 pagineMF1547Front Linkage - Seat PDFAhmad Ali NursahidinNessuna valutazione finora
- Sadi Mohammad Naved: Duties/ResponsibilitiesDocumento3 pagineSadi Mohammad Naved: Duties/ResponsibilitiesNick SanchezNessuna valutazione finora
- AeDocumento12 pagineAeRoberto SanchezNessuna valutazione finora
- Ladybug5 DatasheetDocumento2 pagineLadybug5 DatasheetEloy Ricardo Franco FigueiraNessuna valutazione finora
- Effect of CSR on Corporate Reputation and PerformanceDocumento13 pagineEffect of CSR on Corporate Reputation and PerformanceAnthon AqNessuna valutazione finora
- MS Gree Aircon Midwall Service Manual PDFDocumento55 pagineMS Gree Aircon Midwall Service Manual PDFMacSparesNessuna valutazione finora
- Checklist For BrickworkDocumento2 pagineChecklist For Brickworkइंजि कौस्तुभ पवारNessuna valutazione finora
- 12 585 Rov Latches Brochure v3 Web 1 PDFDocumento8 pagine12 585 Rov Latches Brochure v3 Web 1 PDFIZayvenkoNessuna valutazione finora
- SwephprgDocumento94 pagineSwephprgAbhisekAcharyaNessuna valutazione finora
- Tata Safari 2.2L Headlamp Level Adjustment GuideDocumento3 pagineTata Safari 2.2L Headlamp Level Adjustment GuidePraveen SinghNessuna valutazione finora
- Spare Parts List: Hydraulic Breakers RX6Documento16 pagineSpare Parts List: Hydraulic Breakers RX6Sales AydinkayaNessuna valutazione finora
- Design of Coin Sorter Counter Based On MCU: Articles You May Be Interested inDocumento5 pagineDesign of Coin Sorter Counter Based On MCU: Articles You May Be Interested inArchana BenkarNessuna valutazione finora
- LRP I Approved ProjectsDocumento1 paginaLRP I Approved ProjectsTheReviewNessuna valutazione finora
- Serrano y Olalla - Linearization of The Hoek y BrownDocumento11 pagineSerrano y Olalla - Linearization of The Hoek y BrownEsteban JamettNessuna valutazione finora
- Datasheet Modevapac v2.1 PDFDocumento4 pagineDatasheet Modevapac v2.1 PDFParvezNessuna valutazione finora
- Super Capacitors ConstructionDocumento7 pagineSuper Capacitors ConstructionVivek BavdhaneNessuna valutazione finora
- CIRCULAR WATER TANK DESIGN-Layout1 AkhilDocumento1 paginaCIRCULAR WATER TANK DESIGN-Layout1 AkhilVENKAT KALYANNessuna valutazione finora
- Reliability and Failure Modes of A Hybrid Ceramic Abutment PrototypeDocumento5 pagineReliability and Failure Modes of A Hybrid Ceramic Abutment PrototypeAli FaridiNessuna valutazione finora
- A Young Lasallian Is US Environmental ScholarDocumento1 paginaA Young Lasallian Is US Environmental ScholarDia DimayugaNessuna valutazione finora
- Asi X Packer 105243965Documento3 pagineAsi X Packer 105243965Esteban RochaNessuna valutazione finora
- SUMMER TRAINING REPORT AT Elin Electronics Ltd. Gzb.Documento54 pagineSUMMER TRAINING REPORT AT Elin Electronics Ltd. Gzb.Ravi Kumar100% (3)
- IECDocumento52 pagineIECM.r. Munish100% (2)
- Spokane County Sheriff's Internal Communication PlanDocumento11 pagineSpokane County Sheriff's Internal Communication Planjmcgrath208100% (1)
- Manufacturing Technology Question Papers of JntuaDocumento15 pagineManufacturing Technology Question Papers of JntuaHimadhar SaduNessuna valutazione finora
- PQA824 ManualDocumento100 paginePQA824 ManualElkin AguasNessuna valutazione finora
- Model 2000 Flow ComputerDocumento8 pagineModel 2000 Flow ComputerAdnan SalihbegovicNessuna valutazione finora
- User's Manual: Motherboard AMD Socket AM2Documento56 pagineUser's Manual: Motherboard AMD Socket AM2vagnerrock007Nessuna valutazione finora
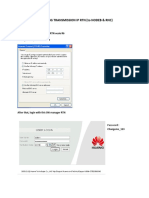
- How to ping NodeB and RNC using RTN transmission IPDocumento14 pagineHow to ping NodeB and RNC using RTN transmission IPPaulo DembiNessuna valutazione finora
- 30x173 - TPDS-T - MK317 Mod0 - NAVSEA - 2011Documento16 pagine30x173 - TPDS-T - MK317 Mod0 - NAVSEA - 2011Anonymous jIzz7woS6Nessuna valutazione finora