Documenti di Didattica
Documenti di Professioni
Documenti di Cultura
Chem 40 Enzyme Kinetics
Caricato da
Justine Grace Mariano100%(1)Il 100% ha trovato utile questo documento (1 voto)
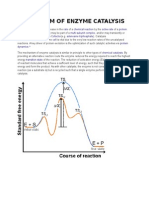
214 visualizzazioni85 pagineEnzymes are catalysts that allow biological reactions to occur at a faster rate. Common names for enzymes are usually formed by adding the suffix -ase to the name of the reactant. A true catalyst increases the rate of reaction by lowering the activation energy barrier.
Descrizione originale:
Copyright
© Attribution Non-Commercial (BY-NC)
Formati disponibili
PDF, TXT o leggi online da Scribd
Condividi questo documento
Condividi o incorpora il documento
Hai trovato utile questo documento?
Questo contenuto è inappropriato?
Segnala questo documentoEnzymes are catalysts that allow biological reactions to occur at a faster rate. Common names for enzymes are usually formed by adding the suffix -ase to the name of the reactant. A true catalyst increases the rate of reaction by lowering the activation energy barrier.
Copyright:
Attribution Non-Commercial (BY-NC)
Formati disponibili
Scarica in formato PDF, TXT o leggi online su Scribd
100%(1)Il 100% ha trovato utile questo documento (1 voto)
214 visualizzazioni85 pagineChem 40 Enzyme Kinetics
Caricato da
Justine Grace MarianoEnzymes are catalysts that allow biological reactions to occur at a faster rate. Common names for enzymes are usually formed by adding the suffix -ase to the name of the reactant. A true catalyst increases the rate of reaction by lowering the activation energy barrier.
Copyright:
Attribution Non-Commercial (BY-NC)
Formati disponibili
Scarica in formato PDF, TXT o leggi online su Scribd
Sei sulla pagina 1di 85
Catalysis
Enzymes catalysts that allow biological reactions to
occur at a faster rate
Ea energy input required to initiate the reaction
Properties of A True Catalyst
Increases the rate of reaction by lowering the
activation energy barrier
Not used up or permanently changed during the
catalytic process
Does not change the position of equilibrium, only the
rate at which equilibrium is attained
Usually acts by forming a transient complex with the
reactant, thus stabilizing the transition state
Enzyme
Cofactor may be an organic or organometallic
molecule (coenzyme) or a metal ion ( Zn
2+
, Mg
2+
,
Cu
2+
)
Holoenzyme complete molecular package, protein
and cofactor
Apoenzyme protein component
Naming Enzyme
Common names for enzymes are usually formed by
adding the suffix ase to the name of the reactant
Tyrosinase catalyzes the oxidation of tyrosine
Cellulase catalyzes the hydrolysis of cellulose to produce
glucose
Some early enzyme names are less descriptive and
give no clue of their function or substrates
Trypsin, chymotrypsin
Enzyme nomenclature
6 major classes, each w/ subsclasses according to
rxn catalyzed
Each is assigned with a 4-digit number and
systematic name which identifies the rxn catalyzed.
Enzyme nomenclature
4-digit official number
1
st
digit: class name
2
nd
digit: subclass
3
rd
digit: accepting functional group
4
th
digit: accepting molecule
EC 1 Oxidoreductases
EC 1.1 Acting on CH-OH group of donors
EC 1.2 Acting on aldehyde or oxo group of
donors
EC 1.3 Acting on CH-CH group of donors
EC 1.4 Acting on CH-NH2 group of donors
EC 1.5 Acting on CH-NH group of donors
EC 1.6 Acting on NADH or NADPH
Kinetics
In a reaction of the form A + B P, the rate can be
expressed in terms of the rate of disappearance of
one of the reactants or in terms of the rate of
appearance of the product
Rate of disappearance of A = -A[A]/At where A symbolizes the
change on [A] and t is time
Rate of appearance of P = A[P]/At
So we can express rate in terms of any of these :
Rate = -A[A]/At = -A[B]/At = A[P]/At
Kinetics
It is established that Rate o [A]
f
[B]
g
Rate = k [A]
f
[B]
g
Where k is the rate constant (proportionality factor that accounts
for how well the reacting molecules oriented during collisions)
Exponents f and g usually small whole numbers 1 or 2 or in some
cases 0 must be determined experimentally
The overall order of the reaction is the sum of all the
exponents
Rate = k[A]
1
first order reaction w/ respect to A and first
order overall
Kinetics
First order reaction rate depends only on one
reactant
Second order reaction the rate of reaction depends
on the two reactants
Rate = k[A]
1
[B]
1
first order wrt A and first order wrt B and
second order overall
Zero order reaction rate is constant depends not on
concentrations of reactants but on other factors
(presence of catalyst)
A B, Rate = k[A]
0
Kinetic Properties of Enzymes
Initial rate (u
0
) determined during the first few
minutes of the reaction
Maximum velocity
(V
max
)
Michaelis-Menten Equation
Leonor Michaelis and Maud Menten explain the
hyperbolic rate curve
Proposed that enzyme molecules, E, and substrate molecules,
S, combine in a fast and reversible step to form an ES complex
E + S
k1
k2
ES
k3
k4
E + P where k are rate constants
Assumptions:
1. Neglect the reaction that reverts product P and free enzyme to
the ES complex defined by k4
2. ES complex is a steady-state intermediate
After mixing E and S, a certain level of ES is formed and its
concentration remain constant because it is produced at the
same rate as it breaks down
Time course of S
consumption, P
formation and
establishment of
steady state level of
ES
Bottom curve: detail
of early stage of time
course
M-M equation
] [
] [
max
0
S K
S V
M
+
= v
Km
Km Michaelis constant
In E + S
k1
k2
ES
k3
E + P, Km is expressed as:
Let Km = [S]
] [
] [ max
0
S K
S V
m +
= v
] [ ] [
] [
max
0
S S
S V
+
= v
] [ 2
] [
max
0
S
S V
= v
2
max
0
V
= v
1
3 2
k
k k
Km
+
=
Km
Remember:
Km becomes:
This is equal to the equilibrium constant for the dissociation of
the ES complex
ES
k2
k1
E + S
So when k2>>k3, Km is simply the dissociation
constant for the ES complex
1
3 2
k
k k
Km
+
=
1
2
k
k
Km =
1
2
] [
] ][ [
k
k
ES
S E
Keq = =
Km
Km measure of how tightly the substrate is bound
to the enzyme
The greater the value of Km, the higher is k2 than k1
Km is the inverse measure of the affinity of the
enzyme for the substrate
Vmax and K3
If the substrate concentration is so high so that E is
completely saturated : [ES] = [E
T
] and V= Vmax
From this part of the enzyme equation:
ES
k3
k4
E + P
Turnover number, k3 (kcat or kp) number of moles
of substrate transformed to product per mole of
enzyme in a defined time period
] [
max
3
T E
V
k =
] [ 3 ES k V =
] [ 3 max T E k V =
K
M
, K
cat
Carbonic anhydrase -CO
2
:
K
cat
= 1 x 10
6
s
-1
;
K
M
= 1.2 x 10
-2
M
Carbonic anhydrase -HCO
3
-
:
K
cat
= 4 x 10
5
s
-1
K
M
= 2.6 x 10
-2
M
Which is better substrate for the enzyme?
K
M
, K
cat
Carbonic anhydrase - CO
2
:
K
cat
= 1 x 10
6
s
-1
;
K
M
= 1.2 x 10
-2
M
Carbonic anhydrase - HCO
3
-
K
cat
= 4 x 10
5
s
-1
K
M
= 2.6 x 10
-2
M
K
cat
/K
M
= 8.3 X10
7
M
-1
s
-1
K
cat
/K
M
= 1.5 X10
7
M
-1
s
-1
Lineweaver-Burk Equation
Double reciprocal plot allows one to plot
experimental enzyme rate data in the form of a
straight line
y = m x + b
] [
] [ 1
max
0 S V
S K
M
+
=
v
max
max
0
1
] [
1 1
V S V
K
M
+ =
v
] [
] [
] [
1
max max
0 S V
S
S V
K
M
+ =
v
] [
] [
max
0
S K
S V
M
+
= v
max
max
0
1
] [
1 1
V S V
K
M
+ =
v
Other Methods of Km and Vmax
Determination
Eadie-Hofstee Plot
The Michaelis-Menten equation is rearranged to
This is a graph of u
0
vs u
0
/[S] where:
Slope = -Km
Y-intercept = Vmax
X-intercept = Vmax/Km
max
0
0
] [
V
S
Km + =
v
v
Eadie-Hofstee
Other Methods of Km and Vmax
Determination
Hanes-Woolf Plot
The Michaelis-Menten equation is rearranged to
This is a graph of [S]/u
0
vs [S] where:
Slope = 1/Vmax
Y-intercept = Km/Vmax
X-intercept = -Km
max max 0
] [
1 ] [
V
Km
S
V
S
+ =
v
Hanes-Woolf
Characteristics of Enzyme Reactions
1. Enzyme
2. pH enzymes have an optimal pH at which they
function most effectively (pH 6-8)
Characteristics of Enzyme Reactions
1. Enzyme
2. pH enzymes have an optimal pH at which they
function most effectively (pH 6-8)
3. Temperature enzymes are sensitive to
temperature changes
For most enzymes, the rate decline begins in the temp.
range of 50C to 60 C
Binding of Substrate to Enzyme
Active site specific region in the enzyme where the
substrate specifically binds
Pocket or crevice in the 3-D structure of the enzyme
Consists of certain amino acids that may be involved with the
noncovalent interaction with the substrate
Characteristics of The Active Site
1. Specificity it is able to discriminate among
possible substrate molecules
Two Types
1. Absolute accept only one type of molecule (can even
discriminate between a D or L isomer)
2. Group Specificity accept a number of closely related
substances as long as the reactive functional group is
present
Characteristics of The Active Site
2. Relatively small, 3-D region within the enzyme aa
residues need not be contiguous in the linear
protein chain
3. Holds substrate through weak, noncovalent,
reversible interactions hydorphobic, ionic and H-
bonding
First Step
Binding of substrate to the enzyme
Three Models
Lock and key model
Induced-fit model
Transition-state model
Lock-and-Key model
Induce-fit model
Transition state model
Increase in Reaction Rate
1. Entropy loss in the ES formation
2. Destabilization of ES due to strain, desolvation or
electrostatic effect
Loss of Entropy
AG = AH - TAS
ES complex is highly organized compared to E and S
in solution
E and S have translational entropy (freedom to
move in 3-D) as well as rotational entropy (freedom
to rotate or tumble about in an axis)
Destabilization of ES Complex
By strain or distortion consequence of the fact that
the enzyme is designed to bind the transition state
more strongly than the substrate
By desolvation of charged groups in the substrate
charged groups are highly stabilized in water
By electrostatic destabilization when a substrate
enters the active site, charged groups may be forced
to interact with groups with same charge resulting to
repulsion and destabilization
2
nd
Step
The transition state is formed and catalysis can occur
Proximity and orientation speed up the reaction in
the transition state, the substrate is bound close to
atoms with which it is to react and also placed in the
correct orientation wrt those atoms
Mechanistic Features of Enzymes
General Acid-Base
Catalysis
Step 1 H
+
is added to
the carbonyl group
Step 2 formation of
the tetrahedral
intermediate
Step 3 a proton is
transferred from O to
N
Step 4 a base will
assist by accepting the
proton from the
intermediate
Mechanistic Features of Enzymes
Metal-ion Catalysis alkali metal ions (Na+, K+)
and transition metals (Mg
2+,
Mn
2+,
Cu
2+
, Zn
2+
,
Fe
2+
,
Fe
3+
,
Ni
2+
,
and others)
1. Holds a substrate properly oriented by coordinate
covalent bonds
Mechanistic Features of Enzymes
Metal-ion Catalysis
2. Enhance reaction by polarizing the scissile bond or
by stabilizing a negatively charged intermediate
Mechanistic Features of Enzymes
Metal-ion Catalysis
3. Participate in biological oxidation-reduction
reactions by reversible electron transfer between
metal ions and substrate
Acts as a Lewis acid by accepting electrons
Mechanistic Features of Enzymes
Covalent Catalysis nucleophilic substitution
reaction
A nucleophilic, (electron-rich) functional group attacks an
electron-deficient group
Nucleophile Leaving
group
Active Site Events
Functional groups in the active site that can have
catalytic roles:
Imidazole ring of histidine
Hydorxyl group of serine
Carboxyl side chain of aspartate and glutamate
Sulfhydryl group of cysteine
Amine group of lysine
Phenol group of tyrosine
Mechanism of Chymotrypsin Action
Serine residue at position 195 is required for activity
Another critical aa in chymotrypsin is His 57
Ser 195
His 57
Asp 102
Enzyme Activity Regulation
Irreversible inhibitor forms covalent or very strong
noncovalent interactions with the enzyme
E-H + R-X E-R + HX
(active) (inactive)
Example: aspirin (acetylsalicylic acid)
Acts by blocking synthesis of pain-producing prostaglandins
Reversible Inhibitors
Can readily combine with and dissociate from an
enzyme and render the enzyme inactive only when
bound
EI is held by weak, noncovalent interactions similar
to ES complex
Competitive Inhibition
Inhibitor usually resembles the structure of normal
substrate and is capable of binding to the active site
of the enzyme
Transition state analogs modeled after the
structures of substrate in presumed transition states
Noncompetitive Inhibition
Inhibitor can bind to the enzyme at a site other than
the active site of the enzyme
Binding of inhibitor causes a change in the structure
of the enzyme especially around the active site
Uncompetitive Inhibition
Inhibitor binds only to the ES complex but not with
the free enzyme
Influence the activity of the enzyme only when [S] is
high and, in turn, the ES concentrations are high
Kinetics of Inhibition
Competitive inhibition slope and x-intercept of
Lineweaver-Burk plot change but the y-intercept
does not
Km increase more substrate is needed to get the velocity of
half the maximum velocity
Kinetics of Inhibition
Pure noncompetitive inhibition - slope and y-
intercept of Lineweaver-Burk plot change but the x-
intercept does not
Pure - Vmax decreases but Km remains the same because the
inhibitor does not interfere with the binding of substrate to the
active site
Mixed Vmax decreases and Km increase because the
inhibitor affects the binding of the substrate to the active site
Kinetics of Inhibition
Uncompetitive inhibition Vmax and Km both
decrease
Reversal of inhibition is not achieved by increase in
[S]
Allosteric Enzymes
Concentration of final products (feedback inhibition)
Concentration of the beginning substrate
Concentration of an intermediate formed in the
sequence
Concentration of external factors (hormones)
Effectors
Biomolecules that influence the action of an
allosteric enzyme
Positive effectors stimulants
Negative effectors - inhibitors
Catalytic and Regulatory Sites
Catalytic site where substrate binds
Regulatory site the binding of effector molecules
changes the conformation of the protein in a way
that tells the other subunits that it is bound
Chemical Alterations
1. Phosphorylation of hydroxyl groups of serine,
threonine or tyrosine
2. Attachment of an adenosyl monophosphate to a
hydroxyl group
3. Reduction of cysteine disulfide bonds
Catalyzed by phosphorylase kinase
Catalyzed by phosphorylase kinase
Catalyzed by phosphorylase phosphatase
Glutamine synthetase
Proteolytic Cleavage
Zymogen inactive protein precursor
Cleaved at one or a few specific peptide bonds to produce the
active form of the enzyme
Regulation by Isoenzyme
Isoenzymes multiple forms of enzymes that have
similar but not identical amino acid sequences
May demonstrate the same enzyme activity but the may differ
in kinetics (Km and Vmax), differ in effectors, differ in the
form of coenzyme needed and cellular distribution
Potrebbero piacerti anche
- Channels, Carriers, and Pumps: An Introduction to Membrane TransportDa EverandChannels, Carriers, and Pumps: An Introduction to Membrane TransportNessuna valutazione finora
- Theoretical and Actual CombustionDocumento14 pagineTheoretical and Actual CombustionErma Sulistyo R100% (1)
- Enzyme KineticsDocumento13 pagineEnzyme KineticsalicjadzNessuna valutazione finora
- 1.1 Proteins - Motifs Structural and Functional Domains Protein FamiliesDocumento32 pagine1.1 Proteins - Motifs Structural and Functional Domains Protein FamilieskinjalkaNessuna valutazione finora
- Hybridization 1Documento37 pagineHybridization 1Anzari MuhammadNessuna valutazione finora
- Problem Set - Enzymes From LehningerDocumento11 pagineProblem Set - Enzymes From LehningervioletbrownNessuna valutazione finora
- Amylase Assay 2Documento9 pagineAmylase Assay 2Rahman ImudaNessuna valutazione finora
- Chapter 6 McKee Enzyme KineticsDocumento79 pagineChapter 6 McKee Enzyme KineticsSuwahono, M.PdNessuna valutazione finora
- EnzymesDocumento20 pagineEnzymesDr.P.Natarajan100% (1)
- Protein Structure and Folding LectureDocumento18 pagineProtein Structure and Folding Lecturemaryscribd241Nessuna valutazione finora
- V VI EnzymesDocumento59 pagineV VI Enzymesthamizh555100% (2)
- 5 Enzyme Kinetics-InhibitionDocumento40 pagine5 Enzyme Kinetics-InhibitionJoel SmolanoffNessuna valutazione finora
- Chapter 10 Chemical Kinetics IIDocumento131 pagineChapter 10 Chemical Kinetics IIChicken ChickenNessuna valutazione finora
- FUEL CellDocumento34 pagineFUEL CellSyed R ShohanNessuna valutazione finora
- Enzyme Catalysis-Chapter 7 (Part 1)Documento22 pagineEnzyme Catalysis-Chapter 7 (Part 1)OmSilence2651Nessuna valutazione finora
- Practical BiochemistryDocumento35 paginePractical BiochemistryMockinjay100% (1)
- Factors Affecting Enzyme ActionDocumento18 pagineFactors Affecting Enzyme Actionanon_458882066Nessuna valutazione finora
- MORE Practice For Michaelis-Menten ShitDocumento3 pagineMORE Practice For Michaelis-Menten ShitMalcolm Charles100% (1)
- Gene Expression in Prokaryotes.Documento23 pagineGene Expression in Prokaryotes.M.PRASAD NAIDU100% (1)
- Ammonia PlantDocumento10 pagineAmmonia PlantHemal Patel Sam100% (3)
- Lecture Notes Enzyme 2 Enzyme Kinetics WebDocumento29 pagineLecture Notes Enzyme 2 Enzyme Kinetics WebAldren RebaLdeNessuna valutazione finora
- 7-Bioc431 Enzymes Kinetics S15Documento34 pagine7-Bioc431 Enzymes Kinetics S15Shariq Mansoor KhanNessuna valutazione finora
- CH 3 - ProblemsDocumento6 pagineCH 3 - ProblemsKhris Griffis100% (5)
- BIO307 Lecture 5 (Enzyme Kinetics I)Documento11 pagineBIO307 Lecture 5 (Enzyme Kinetics I)Phenyo Mmereki100% (1)
- Assign#2 Enzyme KineticsDocumento3 pagineAssign#2 Enzyme KineticsKaren Castro100% (2)
- Excel 2007 TutorialDocumento5 pagineExcel 2007 TutorialromelcarvajalNessuna valutazione finora
- Name: Kristine Joy Atos Block: BSN 1-D Practice ProblemsDocumento6 pagineName: Kristine Joy Atos Block: BSN 1-D Practice ProblemsJenz Hope Segui Novela100% (3)
- Enzyme KineticsDocumento3 pagineEnzyme KineticsZeny Naranjo100% (2)
- BIO307 Lecture 7 (Enzyme Kinetics III)Documento11 pagineBIO307 Lecture 7 (Enzyme Kinetics III)Phenyo Mmereki100% (2)
- Problem Set On Enzyme Kinetics - FS - 2012 - 2013Documento2 pagineProblem Set On Enzyme Kinetics - FS - 2012 - 2013Chay Alcantara100% (1)
- Voet - Chapt - 12 Properties of EnzymesDocumento102 pagineVoet - Chapt - 12 Properties of Enzymestelmo flowNessuna valutazione finora
- MIT Lecture 6 5.07 Biochemistry LectureDocumento16 pagineMIT Lecture 6 5.07 Biochemistry LectureakiridoNessuna valutazione finora
- EnzEng 2 EnzymeKinetics C V VIIDocumento43 pagineEnzEng 2 EnzymeKinetics C V VIIEkuino Simanungkalit100% (1)
- Mod3 PDFDocumento65 pagineMod3 PDFRAKISHO WORLD100% (1)
- Properties of Enzyme Inhibition (CH 3, 7)Documento18 pagineProperties of Enzyme Inhibition (CH 3, 7)afaf100% (1)
- Lecture 1Documento30 pagineLecture 1حموده ابراهيم يونسNessuna valutazione finora
- Methods of Protein AnalysisDocumento41 pagineMethods of Protein AnalysisObin MrazNessuna valutazione finora
- BCH 314 Tutorial 1Documento9 pagineBCH 314 Tutorial 1victorNessuna valutazione finora
- 4 Bioenergetics and Oxidative Metabolism IIDocumento3 pagine4 Bioenergetics and Oxidative Metabolism IILinus LiuNessuna valutazione finora
- Question 1 (37 Marks) : Biochemistry 3 BCH 314Documento4 pagineQuestion 1 (37 Marks) : Biochemistry 3 BCH 314victorNessuna valutazione finora
- 3Documento4 pagine3biotech_vidhya100% (1)
- Protein PurificationDocumento7 pagineProtein PurificationArchana BorahNessuna valutazione finora
- CARBOHYDRATESDocumento38 pagineCARBOHYDRATESgulrukh100% (3)
- Question CH06+answer PDFDocumento8 pagineQuestion CH06+answer PDFCris-Anne Juangco III100% (1)
- Orgo Cheat Sheets 08 2019 PDFDocumento34 pagineOrgo Cheat Sheets 08 2019 PDFKobe AcobNessuna valutazione finora
- Mechanism of Enzyme ActionDocumento2 pagineMechanism of Enzyme ActionBlazy InhumangNessuna valutazione finora
- Polymerase Chain ReactionDocumento14 paginePolymerase Chain ReactionDavidMugambi100% (1)
- Ion Exchange ChromatographyDocumento3 pagineIon Exchange ChromatographysherfudeenNessuna valutazione finora
- Bioenergetics and Oxidative PhosphorylationDocumento32 pagineBioenergetics and Oxidative PhosphorylationShimmering MoonNessuna valutazione finora
- Intracellular TransportDocumento66 pagineIntracellular Transportalvitakhoridatul100% (1)
- Prokaryotic Dna ReplicationDocumento53 pagineProkaryotic Dna ReplicationRININessuna valutazione finora
- Metabolism of Ketone BodiesDocumento38 pagineMetabolism of Ketone BodiesJavedNessuna valutazione finora
- Lecture 2Documento19 pagineLecture 2MuhammadFakhriAimi100% (1)
- Bibc102, Metabolic Biochemistry: Gen-Sheng Feng Fall, 2010Documento14 pagineBibc102, Metabolic Biochemistry: Gen-Sheng Feng Fall, 2010cool_trainerNessuna valutazione finora
- Notes - Alkyl Halides and Aryl HalidesDocumento34 pagineNotes - Alkyl Halides and Aryl HalidesDivya MehtaNessuna valutazione finora
- Lecture 9. Chemical BondingDocumento55 pagineLecture 9. Chemical BondingDione Gale NavalNessuna valutazione finora
- Chapter 2 Water: Multiple Choice QuestionsDocumento10 pagineChapter 2 Water: Multiple Choice QuestionsFrank WuNessuna valutazione finora
- Sample ProblemDocumento1 paginaSample ProblemfintastellaNessuna valutazione finora
- 2.2 Transition Metals Substitution Reactions QsDocumento23 pagine2.2 Transition Metals Substitution Reactions QsJesulayomi BolajiNessuna valutazione finora
- Butadiene and Benzene ManufactureDocumento10 pagineButadiene and Benzene ManufactureAnonymous RJkpep7D0rNessuna valutazione finora
- Mumbai Attacks The Real Story Who Was Behind Mumbai Attack Eye Opening Facts About Mumbai AttacksDocumento2 pagineMumbai Attacks The Real Story Who Was Behind Mumbai Attack Eye Opening Facts About Mumbai AttacksUmar MahmoodNessuna valutazione finora
- Mechanism of Enzyme Catalysis MadhuDocumento9 pagineMechanism of Enzyme Catalysis MadhumengelhuNessuna valutazione finora
- MKBS313 CT1 MemoDocumento3 pagineMKBS313 CT1 MemoLeane MinnaarNessuna valutazione finora
- Vinicius Rossa, Production of Solketal Using Acid Zeolites As CatalystsDocumento18 pagineVinicius Rossa, Production of Solketal Using Acid Zeolites As Catalystsnysa arientikaNessuna valutazione finora
- Preparation of A New Precipitated Iron Catalyst F-T - FujimotoDocumento5 paginePreparation of A New Precipitated Iron Catalyst F-T - FujimotoAngélica ForgionnyNessuna valutazione finora
- Statistical Analysis of FCCU Data To Monitor and Optimize Unit PerformanceDocumento4 pagineStatistical Analysis of FCCU Data To Monitor and Optimize Unit Performancesaleh4060Nessuna valutazione finora
- Report 1 - Feasibility Study For Formalin ProductionDocumento51 pagineReport 1 - Feasibility Study For Formalin ProductionGizem Arslan100% (2)
- Kcet - Chemistry - 2019: Version Code: D-5Documento7 pagineKcet - Chemistry - 2019: Version Code: D-5Manoj CNessuna valutazione finora
- Simulation of An Industrial Turbulent Uidized Bed Reactor For Partial Oxidation To Maleic AnhydrideDocumento10 pagineSimulation of An Industrial Turbulent Uidized Bed Reactor For Partial Oxidation To Maleic AnhydrideCatalinaManjarresNessuna valutazione finora
- Fundamentals and Catalytic Applications of CeO2 Based MaterialsDocumento55 pagineFundamentals and Catalytic Applications of CeO2 Based Materialsjiawen dingNessuna valutazione finora
- Applied Clay Science: Robert A. SchoonheydtDocumento6 pagineApplied Clay Science: Robert A. SchoonheydtDan StefanNessuna valutazione finora
- EnzymesDocumento38 pagineEnzymesSomya Mehndiratta100% (1)
- Green DieselDocumento11 pagineGreen DieselYuvia KusumaNessuna valutazione finora
- A.balla Rev - Chim.2015Documento7 pagineA.balla Rev - Chim.2015abderrahimnNessuna valutazione finora
- Chemical Reactor Design: 7 Sem, B.Tech. Chemical EnggDocumento10 pagineChemical Reactor Design: 7 Sem, B.Tech. Chemical EnggMohammad Ajaz DeshmukhNessuna valutazione finora
- 1.5 Enzyme Kinetics 2Documento25 pagine1.5 Enzyme Kinetics 2Astra BeckettNessuna valutazione finora
- 35-3001-12H Hydrogen/Methane Sample-Draw Detector Operator's ManualDocumento23 pagine35-3001-12H Hydrogen/Methane Sample-Draw Detector Operator's ManualpcatruongNessuna valutazione finora
- Intro Bio QuestionsandanswerarchiveDocumento323 pagineIntro Bio Questionsandanswerarchivesannsann100% (1)
- Igcse Chemistry Theory ZNotesDocumento23 pagineIgcse Chemistry Theory ZNotesQais PMNessuna valutazione finora
- SS2 Chemistry (2nd Term)Documento7 pagineSS2 Chemistry (2nd Term)kazosky4topNessuna valutazione finora
- HydroDocumento16 pagineHydroLutfi GunawanNessuna valutazione finora
- New Methanol Synthesis Catalyst From TopsoeDocumento2 pagineNew Methanol Synthesis Catalyst From TopsoesatishchemengNessuna valutazione finora
- United States Patent (19) : Powers Et Al. (11) Patent NumberDocumento11 pagineUnited States Patent (19) : Powers Et Al. (11) Patent NumberridanormaNessuna valutazione finora
- LAB REPORT 3 - Rate of Reaction PDFDocumento14 pagineLAB REPORT 3 - Rate of Reaction PDFCyrine FranciscoNessuna valutazione finora
- KAU Curriculum of ChemistryDocumento47 pagineKAU Curriculum of ChemistrygogookNessuna valutazione finora