Documenti di Didattica
Documenti di Professioni
Documenti di Cultura
Mann - OU Biochemistry Senior Thesis Poster
Caricato da
Test Test2Descrizione originale:
Copyright
Formati disponibili
Condividi questo documento
Condividi o incorpora il documento
Hai trovato utile questo documento?
Questo contenuto è inappropriato?
Segnala questo documentoCopyright:
Formati disponibili
Mann - OU Biochemistry Senior Thesis Poster
Caricato da
Test Test2Copyright:
Formati disponibili
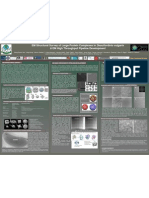
The Wu Lab | Top-down MS and Functional Proteomics
Detection of Protein-Protein Interactions using a 2-Dimensional Chemical Crosslinking Activity Correlated Proteomics
Platform (2D-XL-ACPP)
Morgan Mann | Hongyan Ma | Zhe Wang | Si Wu
Department of Chemistry & Biochemistry, University of Oklahoma, Norman, OK 73019
Overview 2D-ACPP XL-ACPP
Functional protein characterization remains one of the major challenges for Schematic of the 2D-ACPP
proteomics, owing in part, to the significant relationship between protein structure
and function. Current methods to determine higher order protein structure (i.e. X-
Normalized Intensity
Ray Crystallography, NMR, Pull-down etc.) are often difficult and time- Protein 3—Protein 5
consuming—requiring significant resources to isolate relatively few interactions. Protein 3—Protein n
To address this issue, we developed an ACPP that systematically correlates
proteins throughout a chromatographic separation to identify protein-protein Time
interactions. Furthermore, we applied a 2-Dimensional fractionation approach, Protein 1—Protein 2
• Some chromatographic separation methods can disrupt protein interactions and impact correlation.
which leveraged the good orthogonality between Size-Exclusion Chromatography
(SEC) and Anion Exchange Chromatography (AEX) to reduce random matching.
Normalized Intensity
Furthermore, we have begun implementing a Chemical Crosslinking (XL) step to Protein 2—Protein 3
facilitate the use of non-native separation methods with greater resolution.
Time
Hemoglobin 2-Dimensional 2-Dimensional
Background Mixture Fractionation
ACPP
Correlation
• Crosslinking agents —such as Disuccinimidyl suberate (DS))—can covalently link associated
• The 2D-ACPP can reduce the incidence of false-positives by requiring correlation in 2 Dimensions. proteins together to prevent dissociation.
• Proteins mediate almost all physiological and
pathological processes in organisms. • Developed and optimized using a commercially available mixture of Hemoglobin and blood proteins. • Crosslinking products can significantly add to sample complexity.
• e.g. Hemoglobin transport oxygen through
the blood.
Results Results
Chromatographic Orthogonality 2-Dimensional Correlation Crosslinking Efficiency Chromatographic Orthogonality
• Higher-order protein structure is determined
SEC XL AEX XL
by interactions with other proteins, which form SEC AEX SEC: R > 0.90
macromolecular complexes. AEX: R > 0.90
64 KDa
• Hemoglobin is a tetramer of two subunits.
• Protein function is determined by protein 32 KDa
structure. Figure 1. Structure of
Human hemoglobin. [2] 16 KDa
• Hemoglobin transports oxygen significantly Hemoglobin is composed of
more efficiently than monomeric two different protein subunits. Crosslinked
Hemoglobin
equivalents Hemoglobin
• Current methods to detect these interactions are difficult, slow, and require
significant amounts of sample preparation. Figure 4. SDS-PAGE Gel of the Figure 5. UV Chromatograms and SDS-PAGE Gels
Hemoglobin Mixture before and after
• Cannot be applied to biological samples in native-like conditions or helpful Figure 2. UV Chromatograms and SDS-PAGE Figure 3. Protein Interaction candidates
of the crosslinked sample during SEC & AEX
crosslinking. Crosslinked Hemoglobin does separation. Chromatogram peaks are shifted and
timeframes Gels of the Hemoglobin Mixture during both with comparison between separation not present a monomer (~16 KDa), and the broadened relative to the unlinked sample, which is
separation methods (SEC & AEX). Molecular methods. 9 Hemoglobin interactions were intensity of higher molecule bands is
• New, higher throughput methods will aid studies in a variety of fields, ranging consistent with increased sample complexity. SDS-
weight distributions indicate that the separation detected, out of 15 total interactions. 110 significantly increased—indicating that PAGE indicates that the separation methods are
from cancer and disease research to basic biochemistry. methods are orthogonal. proteins were detected and correlated in this presence of crosslinking products. orthogonal.
sample.
Protein ACPP Conclusions Future Directions
1. The ACPP can efficiently identify Hemoglobin proteins in a sample consisting of 1. Application to more complex samples to confirm efficacy in biological samples.
over 110 proteins. • E. coli lysate spiked with Hemoglobin (1:30).
2. The 2D-ACPP significantly reduced the number of non-Hemoglobin matches 2. Use of more powerful separation methods to evaluate the XL-ACPP.
Proteome
Size-Exclusion
Protein Co-Elution Statistical Correlation
detected. • Reverse-Phase Chromatography (RPLC).
Chromatography
3. Disuccinimidyl suberate can efficiently crosslink Hemoglobin. 3. In-vivo crosslinking to insure native conditions during crosslinking.
• Interacting proteins will coelute during separation and correlate well.
4. Size-Exclusion Chromatography and Anion Exchange Chromatography demonstrate 4. Improvements to the Correlation Algorithm
• A correlation coefficient (R-score) Threshold of 0.90 was utilized.
good orthogonality in both unlinked and crosslinked samples.
• Protein Interactions can be identified in a High-Throughput manner.
• Random Coelution can generate false-positives.
Contact Acknowledgements References
Morgan Mann, Undergraduate Research Assistant The authors would like to thank the members of the Wu Laboratory for their constant help and 1. Ma, H.; Delafield, D. G.; Wang, Z.; You, J.; Wu, S., Finding Biomass Degrading Enzymes Through an Activity-Correlated
morgan.w.mann@ou.edu guidance. The authors would also like to thank the University of Oklahoma for providing the Quantitative Proteomics Platform (ACPP). J. Am. Soc. Mass Spectrom. 2017, 28 (4), 655-663
101 Stephenson Parkway, Norman, OK 73019 opportunity to conduct and present this research 2. Zephyris. Hemoglobin. English language Wikipedia. https://commons.wikimedia.org/w/index.php?curid=2300973
Potrebbero piacerti anche
- Bioinformatics Part 3 Protein Interactions Molecular DockingDocumento105 pagineBioinformatics Part 3 Protein Interactions Molecular DockingKhuwaylaNessuna valutazione finora
- Phys. Chem. Chem. Phys. 2016, 18, 24506Documento5 paginePhys. Chem. Chem. Phys. 2016, 18, 24506FARM 1Nessuna valutazione finora
- Chem41290 Part VDocumento13 pagineChem41290 Part VJulia MachajNessuna valutazione finora
- Coarse Grained Lattice Folding QuantumDocumento12 pagineCoarse Grained Lattice Folding QuantumJehangir VakilNessuna valutazione finora
- ABX Pentra Total Protein CPDocumento6 pagineABX Pentra Total Protein CPIbrahimAliNessuna valutazione finora
- Structure - Based Virtual Screening - Case StudiesDocumento22 pagineStructure - Based Virtual Screening - Case Studiesapi-3787547Nessuna valutazione finora
- I-TASSER: A Unified Platform For Automated Protein Structure and Function PredictionDocumento14 pagineI-TASSER: A Unified Platform For Automated Protein Structure and Function PredictionDavidNessuna valutazione finora
- Separation Techniques: Chromatography: November 2016Documento6 pagineSeparation Techniques: Chromatography: November 2016JacKLOPNessuna valutazione finora
- "Physiologically Based Pharmacokinetics Modelling": Yos Adi Prakoso, DVM, MSCDocumento19 pagine"Physiologically Based Pharmacokinetics Modelling": Yos Adi Prakoso, DVM, MSCGerald JoshuaNessuna valutazione finora
- SPR Chemical EducationDocumento7 pagineSPR Chemical EducationAdam TilloNessuna valutazione finora
- An Introduction To Proteomics: The Protein Complement of The GenomeDocumento40 pagineAn Introduction To Proteomics: The Protein Complement of The GenomeJohn Louie BarquerosNessuna valutazione finora
- Acs Analchem 1c02365Documento8 pagineAcs Analchem 1c02365Nicolas ÑustesNessuna valutazione finora
- Riginal Article: Pritam Jain, Amar Chaudhari, Anup Bang, Sanjay SuranaDocumento6 pagineRiginal Article: Pritam Jain, Amar Chaudhari, Anup Bang, Sanjay SuranaAmarNessuna valutazione finora
- Acs JCTC 2c00743Documento10 pagineAcs JCTC 2c00743wangxy940930Nessuna valutazione finora
- Bioinformatiks in MassDocumento15 pagineBioinformatiks in Masstexto.sarlNessuna valutazione finora
- Glyconanoparticles For Targeted Tumor Therapy of Platinum Anticancer DrugDocumento11 pagineGlyconanoparticles For Targeted Tumor Therapy of Platinum Anticancer DrugssNessuna valutazione finora
- An Overview On Molecular Docking PDFDocumento14 pagineAn Overview On Molecular Docking PDFMd MoinulNessuna valutazione finora
- Computational Prediction of Structure, Substrate Binding Mode, Mechanism, and Rate For A Malaria Protease With A Novel Type of Active SiteDocumento8 pagineComputational Prediction of Structure, Substrate Binding Mode, Mechanism, and Rate For A Malaria Protease With A Novel Type of Active SiteRagaNessuna valutazione finora
- Acs Analchem 8b05846Documento7 pagineAcs Analchem 8b05846Astryd ParkNessuna valutazione finora
- Huang, Chi, Chien - 2018 - Size-Exclusion Chromatography Using Reverse-Phase Columns For Protein SeparationDocumento12 pagineHuang, Chi, Chien - 2018 - Size-Exclusion Chromatography Using Reverse-Phase Columns For Protein Separationκ.μ.α «— Brakat»Nessuna valutazione finora
- Contemporary Methodology For Protein Structure Determination Hunkapiller Et Al.,1984Documento8 pagineContemporary Methodology For Protein Structure Determination Hunkapiller Et Al.,1984Andrés Jonathan Cepeda GuerronNessuna valutazione finora
- Marrink Aggre ModelsDocumento9 pagineMarrink Aggre Modelssoumava palitNessuna valutazione finora
- Greenbaum D, Colangelo C, Williams K, Gerstein M. Comparing Protein Abundance and mRNA Expression Levels On A Genomic Scale. Genome Biol 4: 117Documento9 pagineGreenbaum D, Colangelo C, Williams K, Gerstein M. Comparing Protein Abundance and mRNA Expression Levels On A Genomic Scale. Genome Biol 4: 117Sg89Nessuna valutazione finora
- An Overview On Molecular DockingDocumento14 pagineAn Overview On Molecular DockingRiya PremNessuna valutazione finora
- Xóa 2Documento15 pagineXóa 2Rin TohsakaNessuna valutazione finora
- Physical Model of The Genotype-to-Phenotype Map of Proteins: Doi: Subject Areas: Biological PhysicsDocumento15 paginePhysical Model of The Genotype-to-Phenotype Map of Proteins: Doi: Subject Areas: Biological PhysicsMariano UriaNessuna valutazione finora
- Monoclonal Antibodies:: Beyond The PlatformDocumento20 pagineMonoclonal Antibodies:: Beyond The PlatformBalaNessuna valutazione finora
- Proteomics Lab ReportDocumento4 pagineProteomics Lab Reportnursah.aslanNessuna valutazione finora
- Iop JopDocumento10 pagineIop JopAkash SrivastavaNessuna valutazione finora
- In-Gel Digestion For MS of Proteomes.2006.468Documento5 pagineIn-Gel Digestion For MS of Proteomes.2006.468qinhuixing3Nessuna valutazione finora
- 3 Lec Secretory Folding HODocumento11 pagine3 Lec Secretory Folding HOHamud ShmalyNessuna valutazione finora
- Investigating Transport Proteins by Solid State NMRDocumento14 pagineInvestigating Transport Proteins by Solid State NMRAArriiss WizushkiNessuna valutazione finora
- Docking SlidesDocumento42 pagineDocking Slidestalal adlanNessuna valutazione finora
- Proteomic Sample Preparation TechniquesDocumento18 pagineProteomic Sample Preparation TechniquesLe TrungNessuna valutazione finora
- Nature 23 High-Throughput Oligopaint Screen Identifies Druggable 3D Genome RegulatorsDocumento33 pagineNature 23 High-Throughput Oligopaint Screen Identifies Druggable 3D Genome Regulatorslandau1994Nessuna valutazione finora
- Batch Adsorption - Confocal 11Documento9 pagineBatch Adsorption - Confocal 11Al-somuda' Ali Al-smmaniNessuna valutazione finora
- MY Proteomics FinalDocumento56 pagineMY Proteomics FinalSaradha PellatiNessuna valutazione finora
- Paper2 EsmnbrDocumento8 paginePaper2 EsmnbrGANYA U 2022 Batch,PES UniversityNessuna valutazione finora
- Department of Biochemistry. University of Virginia. Chorlottexvil!e. Virginin 22901, US.4Documento9 pagineDepartment of Biochemistry. University of Virginia. Chorlottexvil!e. Virginin 22901, US.4JefersonMatosdeColaresNessuna valutazione finora
- Deviation of Trypsin Activity Using Peptide Conformational ImprintsDocumento14 pagineDeviation of Trypsin Activity Using Peptide Conformational ImprintsMarco Antonio Becerril HernandezNessuna valutazione finora
- Chromatographic Protein Refolding/Renaturation: R 2018 Elsevier Inc. All Rights ReservedDocumento17 pagineChromatographic Protein Refolding/Renaturation: R 2018 Elsevier Inc. All Rights ReservedMai LinhNessuna valutazione finora
- Analisis Item Bio SPM 2003-2019Documento4 pagineAnalisis Item Bio SPM 2003-2019Caryn YeapNessuna valutazione finora
- Proteomics 1Documento73 pagineProteomics 1ShreevarNessuna valutazione finora
- EM Structural Survey of Large Protein Complexes in Desulfovibrio VulgarisDocumento1 paginaEM Structural Survey of Large Protein Complexes in Desulfovibrio Vulgarisapi-77382155Nessuna valutazione finora
- Spectroscopy With Nature-Inspired Genetic 3Documento8 pagineSpectroscopy With Nature-Inspired Genetic 3Sonny BrowningNessuna valutazione finora
- 10 1039@c9an02553gDocumento10 pagine10 1039@c9an02553gLeidy Constanza Villalobos GonzalezNessuna valutazione finora
- Proteins - 2007 - Mintseris - Integrating Statistical Pair Potentials Into Protein Complex PredictionDocumento10 pagineProteins - 2007 - Mintseris - Integrating Statistical Pair Potentials Into Protein Complex PredictionInes MejriNessuna valutazione finora
- Analytical Methods: PaperDocumento8 pagineAnalytical Methods: Paperchikh MELKAOUINessuna valutazione finora
- COSTBI 2018 196 Accepted RefcorrectedDocumento14 pagineCOSTBI 2018 196 Accepted RefcorrectedShatanik MukherjeeNessuna valutazione finora
- Workshop Protein Modeling PDFDocumento54 pagineWorkshop Protein Modeling PDFrima rasidaNessuna valutazione finora
- 2019gakiya Teruya SilvernanoparticlesynthesisDocumento6 pagine2019gakiya Teruya Silvernanoparticlesynthesisعبدالعزيز السكافيNessuna valutazione finora
- The in Uence of Residence Time Distribution On Continuous-Flow PolymerizationDocumento7 pagineThe in Uence of Residence Time Distribution On Continuous-Flow PolymerizationNorma JenarezNessuna valutazione finora
- Ref 3 Single Microgels Jidheden 2016Documento13 pagineRef 3 Single Microgels Jidheden 2016ณพดนัย จักรภีร์ศิริสุขNessuna valutazione finora
- Quantitative Mass Spectrometry in Proteomics: A Critical ReviewDocumento15 pagineQuantitative Mass Spectrometry in Proteomics: A Critical Reviewaselle kellyNessuna valutazione finora
- Sabzevari 2018Documento10 pagineSabzevari 2018Abdullah ZndNessuna valutazione finora
- X Ray Crystallography and NMR TechniqueDocumento30 pagineX Ray Crystallography and NMR TechniqueSaran.S.MenonNessuna valutazione finora
- Du Et Al 2024 Synthesis of The Cyclopentane Core Skeleton of Cranomycin and JogyamycinDocumento4 pagineDu Et Al 2024 Synthesis of The Cyclopentane Core Skeleton of Cranomycin and JogyamycinkarthikNessuna valutazione finora
- Muito Detalhada Essa Revisão TranscriptomicaDocumento14 pagineMuito Detalhada Essa Revisão TranscriptomicaLeticia PontesNessuna valutazione finora
- Science 1124619Documento7 pagineScience 11246199868838836ankNessuna valutazione finora
- Utilizing Web-Based Search Engines for Analyzing Biological MacromoleculesDa EverandUtilizing Web-Based Search Engines for Analyzing Biological MacromoleculesNessuna valutazione finora
- Robbins Pathology - Chapter 1 TransDocumento11 pagineRobbins Pathology - Chapter 1 Transnath nathNessuna valutazione finora
- ErbB HER Protein-Tyrosine Kinases Structures and SmallDocumento18 pagineErbB HER Protein-Tyrosine Kinases Structures and SmallprototyposNessuna valutazione finora
- Glucose Levels Maintaining MechanicsDocumento7 pagineGlucose Levels Maintaining MechanicsSukhdeep KumarNessuna valutazione finora
- Encyclopediabookchapter PDFDocumento13 pagineEncyclopediabookchapter PDFJamesNessuna valutazione finora
- Enzymes WorksheetDocumento2 pagineEnzymes WorksheetNatalie Pemberton100% (1)
- NEET MCQ Revision-1 by Tulip AcademyDocumento27 pagineNEET MCQ Revision-1 by Tulip AcademyTulip AcademyNessuna valutazione finora
- Berg 8e Testbank Chapter03Documento8 pagineBerg 8e Testbank Chapter03jsw4117Nessuna valutazione finora
- 16) Signaling Pathways That Control Gene ActivityDocumento27 pagine16) Signaling Pathways That Control Gene ActivityRNessuna valutazione finora
- Protein StructureDocumento10 pagineProtein StructureSzekeres-Csiki KatalinNessuna valutazione finora
- Chromatin and Chromosomes: by Benjamin LewinDocumento46 pagineChromatin and Chromosomes: by Benjamin LewinKavisa GhoshNessuna valutazione finora
- #NEET Fat Soluble VitaminDocumento1 pagina#NEET Fat Soluble VitaminSatvik PatelNessuna valutazione finora
- Growth Factors VS CytokinesDocumento65 pagineGrowth Factors VS CytokinesmahaNessuna valutazione finora
- Komponen Bioaktif Protein Dan Lemak Dalam Susu Kuda LiarDocumento9 pagineKomponen Bioaktif Protein Dan Lemak Dalam Susu Kuda LiarThenMust Andy PrasetioNessuna valutazione finora
- Notes Carb MetabolismDocumento7 pagineNotes Carb MetabolismCrimson MupasNessuna valutazione finora
- Sila Jawab SEMUA Soalan. Semua Jawapan Hendaklah Ditaip, Dilukis Secara Digital (Kecuali Gambar Yang Diminta) Dan Dihantar Dalam Format PDFDocumento7 pagineSila Jawab SEMUA Soalan. Semua Jawapan Hendaklah Ditaip, Dilukis Secara Digital (Kecuali Gambar Yang Diminta) Dan Dihantar Dalam Format PDFJegathiswary GanasanNessuna valutazione finora
- Lipid Lowering AgentsDocumento56 pagineLipid Lowering AgentsDUEÑAS, MARIELNessuna valutazione finora
- Lodish6e ch07Documento8 pagineLodish6e ch07nNessuna valutazione finora
- Walnut Handler List - California WalnutsDocumento7 pagineWalnut Handler List - California WalnutsdaniabiomassNessuna valutazione finora
- Titin ProteinDocumento40 pagineTitin ProteinAlexanderNessuna valutazione finora
- Molecular Biology of The Cell, Sixth Edition Chapter 16: The CytoskeletonDocumento37 pagineMolecular Biology of The Cell, Sixth Edition Chapter 16: The CytoskeletonJeanPaule Joumaa100% (1)
- Bio Inorganic ChemistryDocumento106 pagineBio Inorganic ChemistryUmendra Kumar KhokharNessuna valutazione finora
- Metabolism of Non Glucose SugarsDocumento21 pagineMetabolism of Non Glucose SugarsCLEMENT100% (5)
- Appendix I: IA IUPAC Nucleotide Ambiguity CodesDocumento2 pagineAppendix I: IA IUPAC Nucleotide Ambiguity Codespeeps007Nessuna valutazione finora
- Franco 2017Documento12 pagineFranco 2017ReshmaaRajendranNessuna valutazione finora
- STRA65Documento6 pagineSTRA65SUJITH232323Nessuna valutazione finora
- Enzyme and Acid - Base CatalysisDocumento64 pagineEnzyme and Acid - Base Catalysisbinseung skzNessuna valutazione finora
- Lipid ProfileDocumento23 pagineLipid Profilekyawswakyawswa100% (1)
- Lipid Profile Disease and DiagnosisDocumento31 pagineLipid Profile Disease and DiagnosisGeetanjali Jha100% (1)
- Lesson 8 Biological MoleculeDocumento27 pagineLesson 8 Biological MoleculeChris John RebustesNessuna valutazione finora
- Heteroglycans: Proteoglycans (Gags), Peptidoglycans and GlycoproteinsDocumento13 pagineHeteroglycans: Proteoglycans (Gags), Peptidoglycans and GlycoproteinsSaeed AbdulhadiNessuna valutazione finora