Documenti di Didattica
Documenti di Professioni
Documenti di Cultura
Biochemistry
Caricato da
Loida SarmientoCopyright
Formati disponibili
Condividi questo documento
Condividi o incorpora il documento
Hai trovato utile questo documento?
Questo contenuto è inappropriato?
Segnala questo documentoCopyright:
Formati disponibili
Biochemistry
Caricato da
Loida SarmientoCopyright:
Formati disponibili
Irish Jane Ico DMD 1-A
Dr. Barcarse
Mass Spectrophotometry Over the past decade, mass spectrometry has undergone tremendous technological improvements allowing for its application to proteins, peptides, carbohydrates, DNA, drugs, and many other biologically relevant molecules. A mass spectrometer determines the mass of a molecule by measuring the mass-to-charge ratio (m/z) of its ion. Ions are generated by inducing either the loss or gain of a charge from a neutral species. Once formed, ions are electrostatically directed into a mass analyzer where they are separated according to m/z and finally detected. The result of molecular ionization, ion separation, and ion detection is a spectrum that can provide molecular mass and even structural information.
Nuclear magnetic resonance spectroscopy, most commonly known as NMR spectroscopy, is a research technique that exploits the magnetic properties of certainatomic nuclei to determine physical and chemical properties of atoms or the moleculesin which they are contained. It relies on the phenomenon of nuclear magnetic resonance and can provide detailed information about the structure, dynamics, reaction state, and chemical environment of molecules. Most frequently, NMR spectroscopy is used by chemists and biochemists to investigate the properties of organic molecules, though it is applicable to any kind of sample that contains nuclei possessing spin. Suitable samples range from small compounds analyzed with 1-dimensional proton or carbon-13 NMR spectroscopy to large proteins or nucleic acids using 3 or 4-dimensional techniques. The impact of NMR spectroscopy on the sciences has been substantial because of the range of information and the diversity of samples, including solutions and solids.
High-performance liquid chromatography (sometimes referred to as high-pressure liquid chromatography), HPLC, is a chromatographic technique used to separate a mixture of compounds in analytical chemistry and biochemistry with the purpose of identifying, quantifying and purifying the individual components of the mixture. Some common examples are the separation and quantitation of performance enhancement drugs (e.g. steroids) in urine samples, or of vitamin D levels in serum. HPLC typically utilizes different types of stationary phases (i.e. sorbents) contained in columns, a pump that moves the mobile phase and sample components through the column, and a detector capable of providing characteristic retention times for the sample components and area counts reflecting the amount of each analyte passing through the detector. The detector may also provide additional information related to the analyte, (i.e.UV/Vis spectroscopic data, if so equipped). Analyte retention time varies depending on the strength of its interactions with the stationary phase, the composition and flow rate of mobile phase used, and on the column dimensions. HPLC is a form of liquid chromatography that utilizes small size columns (typically 250 mm or shorter and 4.6 mm i.d. or smaller; packed with smaller particles), and higher mobile phase pressures compared to ordinary liquid chromatography. With HPLC, a pump (rather than gravity) provides the higher pressure required to move the mobile phase and sample components through the densely packed column. The increased density arises from the use of smaller sorbent particles. Such particles are capable of providing better separation on columns of shorter length when compared to ordinary column chromatography.
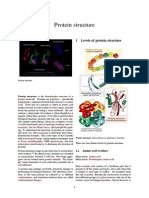
Four Levels of Architectural Organization
Primary structure The primary structure refers to amino acid linear sequence of the polypeptide chain. The primary structure is held together by covalent or peptide bonds, which are made during the process of protein biosynthesis or translation. The two ends of thepolypeptide chain are referred to as the carboxyl terminus (C-terminus) and the amino terminus (N-terminus) based on the nature of the free group on each extremity. Counting of residues always starts at the N-terminal end (NH2-group), which is the end where the amino group is not involved in a peptide bond. The primary structure of a protein is determined by the gene corresponding to the protein. A specific sequence ofnucleotides in DNA is transcribed into mRNA, which is read by the ribosome in a process called translation. The sequence of a protein is unique to that protein, and defines the structure and function of the protein. The sequence of a protein can be determined by methods such as Edman degradation or tandem mass spectrometry. Often however, it is read directly from the sequence of the gene using the genetic code. We know that there are over 10,000 proteins in our body which are composed of different arrangements of 20 types of amino acid residues(it is strictly recommended to use the word "amino acid residues" as when peptide bond is formed a water molecule is lost so, protein is made up of amino acid residues) Post-translational modifications such as disulfide formation, phosphorylations and glycosylations are usually also considered a part of the primary structure, and cannot be read from the gene. Amino acid residues Each -amino acid consists of a backbone part that is present in all the amino acid types, and a side chain that is unique to each type of residue. An exception from this rule is proline. Because the carbon atom is bound to four different groups it is chiral, however only one of the isomers occur in biological proteins. Glycine however, is not chiral since its side chain is a hydrogen atom. A simplemnemonic for correct L-form is "CORN": when the C atom is viewed with the H in front, the residues read "CO-R-N" in a clockwise direction.
Secondary structure Secondary structure refers to highly regular local sub-structures. Two main types of secondary structure, thealpha helix and the beta strand or beta sheets, were suggested in 1951 by Linus Pauling and coworkers. These secondary structures are defined by patterns of hydrogen bonds between the main-chain peptide groups. They have a regular geometry, being constrained to specific values of the dihedral angles and on the Ramachandran plot. Both the alpha helix and the beta-sheet represent a way of saturating all the hydrogen bond donors and acceptors in the peptide backbone. Some parts of the protein are ordered but do not form any regular structures. They should not be confused with random coil, an unfolded polypeptide chain lacking any fixed three-dimensional structure. Several sequential secondary structures may form a "supersecondary unit". Tertiary structure Tertiary structure refers to three-dimensional structure of a single protein molecule. The alpha-helices and betasheets are folded into a compact globule. The folding is driven by the non-specific hydrophobic interactions (the burial of hydrophobic residues from water), but the structure is stable only when the parts of a protein domain are locked into place by specific tertiary interactions, such as salt bridges, hydrogen bonds, and the tight packing of side chains and disulfide bonds. The disulfide bonds are extremely rare in cytosolic proteins, since the cytosol is generally a reducing environment.
Quaternary structure Quaternary structure is the three-dimensional structure of a multi-subunit protein and how the subunits fit together. In this context, the quaternary structure is stabilized by the same non-covalent interactions and disulfide bonds as the tertiary structure. Complexes of two or more polypeptides (i.e. multiple subunits) are called multimers. Specifically it would be called a dimer if it contains two subunits, a trimer if it contains three subunits, and a tetramer if it contains four subunits. The subunits are frequently related to one another bysymmetry operations, such as a 2-fold axis in a dimer. Multimers made up of identical subunits are referred to with a prefix of "homo-" (e.g. a homotetramer) and those made up of different subunits are referred to with a prefix of "hetero-" (e.g. a heterotetramer, such as the two alpha and two beta chains of hemoglobin).
Tests for Amino Acids and Protein
Quantitative test 1. Spectrophotometric assay Protein shows maximum absorbance at 280nm due to presence of tyrosine and tryptophane. 2. Biuret test shows 540nm 3. Lowry test shows 750nm Qualitative test Ninhydrin Test Amino acid containing a free amino group and a free carboxylic acid group that react together with ninhydrin to produce colure product. Protein also contain free amino group on the alpha-carbon and can react with ninhydrin to produce blue purple product. When the amino group is secondary the condensation product is yellow. Biuret Test It positively identifies the presence of protein in solution with a deep violet colour.Biuret H2NCONHCONH2 reacts with copper(11)ions in a basic solution to form a deep voilet complex. Xanthoproteic Test The amino acids that contain benzene ring like tyrosine and tryptophan undergo nitration in this test and gives yellow colour. Millons Test It is specific for tyrosine, the only amino acid that contain a phenol group on which a hydroxyl group is attached. It gives red precipitate. Hopkins_cole Test It is specific for tryptophan, the only amino acid containing an indole group.The indole ring reacts with glyoxylic acid in the presence of a strong acid to form a violet cyclic product. Nitroprusside Test It is specific for Cysteine, the only amino acid containing sulfahydrylgroup. Forms red complex.Classification and Nomenclature of
Potrebbero piacerti anche
- Protein Structure Notes - Level 1 - Medical StudentsDocumento16 pagineProtein Structure Notes - Level 1 - Medical StudentssafemindNessuna valutazione finora
- Redox Chemistry and Biology of ThiolsDa EverandRedox Chemistry and Biology of ThiolsBeatriz AlvarezNessuna valutazione finora
- Molecular Protien StuctureDocumento5 pagineMolecular Protien Stucturechetan0047Nessuna valutazione finora
- Tetrahedron Reports on Organic Chemistry: Volume 1.1-10Da EverandTetrahedron Reports on Organic Chemistry: Volume 1.1-10Derek BartonNessuna valutazione finora
- Module 5. Proteins Course Outcomes: at The End of The Course, The Student Shall Be Able ToDocumento6 pagineModule 5. Proteins Course Outcomes: at The End of The Course, The Student Shall Be Able ToAldine MabulacNessuna valutazione finora
- Screenshot 2022-01-07 at 1.34.05 PMDocumento25 pagineScreenshot 2022-01-07 at 1.34.05 PMPaolaNessuna valutazione finora
- Biology Lab - Biuret TestDocumento7 pagineBiology Lab - Biuret TestZoe Bradshaw0% (1)
- Full Lab Report On: Exercise No. 4 Protein DenaturationDocumento8 pagineFull Lab Report On: Exercise No. 4 Protein DenaturationElaine FaloNessuna valutazione finora
- Chapter 2 - Protein Structure: B: Probing Composition, Sequence, and ConformationDocumento26 pagineChapter 2 - Protein Structure: B: Probing Composition, Sequence, and ConformationMona ElsayedNessuna valutazione finora
- Week 2 PPT (Laboratory) BiochemDocumento16 pagineWeek 2 PPT (Laboratory) BiochemHumphrey SubitoNessuna valutazione finora
- Proteins & Proteins Purification TechniquesDocumento54 pagineProteins & Proteins Purification Techniquesnurul farhanahNessuna valutazione finora
- Protein Structure PDFDocumento6 pagineProtein Structure PDFmradu1Nessuna valutazione finora
- Protein Levels of Structural Organization in Proteins Primary StructureDocumento19 pagineProtein Levels of Structural Organization in Proteins Primary StructureArisekola OlubodunNessuna valutazione finora
- Biophysics and Structural Bioinformatics IDocumento34 pagineBiophysics and Structural Bioinformatics IMaddy IlNessuna valutazione finora
- Proteins: Fundamental Chemical Properties: Alain J CozzoneDocumento10 pagineProteins: Fundamental Chemical Properties: Alain J CozzoneMarta SilvaNessuna valutazione finora
- Peptides and Proteins: M.Prasad Naidu MSC Medical Biochemistry, PH.DDocumento30 paginePeptides and Proteins: M.Prasad Naidu MSC Medical Biochemistry, PH.DDr. M. Prasad NaiduNessuna valutazione finora
- NOTES Tutorial 4 Proteins RevisionNotes BMSC2120Documento5 pagineNOTES Tutorial 4 Proteins RevisionNotes BMSC2120ZAFFAR QURESHINessuna valutazione finora
- Zarmeena#07Documento15 pagineZarmeena#07Usman AliNessuna valutazione finora
- Proteins (: (Hide) 1 o 1.1 2 o 2.1 o 2.2 3 o 3.1 4 o 4.1Documento4 pagineProteins (: (Hide) 1 o 1.1 2 o 2.1 o 2.2 3 o 3.1 4 o 4.1Satheesh KumarNessuna valutazione finora
- Amino Acids - 2 - NoDocumento9 pagineAmino Acids - 2 - NoSunny Thakur17Nessuna valutazione finora
- Unit I Protein StructureDocumento66 pagineUnit I Protein StructurenikteshgNessuna valutazione finora
- Surface Ftir Techniques To Analyze The Conformation of Proteinspeptides in H2o Environment 2161 0398 1000202Documento8 pagineSurface Ftir Techniques To Analyze The Conformation of Proteinspeptides in H2o Environment 2161 0398 1000202S MajumderNessuna valutazione finora
- Bion Peptides and Proteins First Semester 2021 2022Documento55 pagineBion Peptides and Proteins First Semester 2021 2022Hashem Bani yaseenNessuna valutazione finora
- Journal of Molecular Structure: Daojin Li, Yumin Yang, Xinxiang Cao, Chen Xu, Baoming JiDocumento11 pagineJournal of Molecular Structure: Daojin Li, Yumin Yang, Xinxiang Cao, Chen Xu, Baoming JiIoanaCarlanNessuna valutazione finora
- Key Suggestions For MBBS StudentsDocumento14 pagineKey Suggestions For MBBS Studentsnihadabj2003Nessuna valutazione finora
- 02 BCH101 Lecture 2 ProteinDocumento37 pagine02 BCH101 Lecture 2 Proteinsharkar1059Nessuna valutazione finora
- LO Week 4Documento11 pagineLO Week 4Matthew ChristopherNessuna valutazione finora
- Biology NotesDocumento42 pagineBiology NotesJoel ThompsonNessuna valutazione finora
- Proteins: Structure & FunctionsDocumento15 pagineProteins: Structure & FunctionsAbhinav KumarNessuna valutazione finora
- Protein PDFDocumento15 pagineProtein PDFmradu1Nessuna valutazione finora
- Protein (Structures and Functions)Documento46 pagineProtein (Structures and Functions)Dian AgustiarNessuna valutazione finora
- Amino Acids and ProteinsDocumento7 pagineAmino Acids and Proteinslcassidy9074Nessuna valutazione finora
- Protein FunctionDocumento39 pagineProtein FunctionDeana Namirembe100% (1)
- Proteins Include A Diversity of Structures, Resulting in A Wide Range of FunctionsDocumento12 pagineProteins Include A Diversity of Structures, Resulting in A Wide Range of FunctionsDanielle GuerraNessuna valutazione finora
- PKU 2. Mohammad Afifudin A 18030194020 PKO ProteinDocumento9 paginePKU 2. Mohammad Afifudin A 18030194020 PKO Proteinafif armadaniNessuna valutazione finora
- Proteins ChemistryDocumento10 pagineProteins ChemistryDarshan RNessuna valutazione finora
- Structure and Function of Macromolecules: 2.2.1 Carbohydrates-The Energy GiversDocumento12 pagineStructure and Function of Macromolecules: 2.2.1 Carbohydrates-The Energy GiversSteven BanksNessuna valutazione finora
- البروتيناتDocumento54 pagineالبروتيناتYaman HassanNessuna valutazione finora
- Kami Export - ProteinsDocumento33 pagineKami Export - Proteinskrissanya.scampbell1021Nessuna valutazione finora
- Application of 2 D-HPLC System For Plasma Protein SeparationDocumento10 pagineApplication of 2 D-HPLC System For Plasma Protein Separationtrinhmy81Nessuna valutazione finora
- B1. Amino Acid Analysis and Chemical SequencingDocumento4 pagineB1. Amino Acid Analysis and Chemical SequencinganiNessuna valutazione finora
- Biochemistry Experiment JournalDocumento46 pagineBiochemistry Experiment JournalEra MelaniaNessuna valutazione finora
- Proteins Are Good For HealthDocumento30 pagineProteins Are Good For HealthMohsin SheikhNessuna valutazione finora
- Biology A Level SpecDocumento32 pagineBiology A Level SpecszyzypNessuna valutazione finora
- Chemistry of LifeDocumento119 pagineChemistry of LifeRamazan AshirkhanNessuna valutazione finora
- Purification of Protiens: Mohan CC M.SC, Biochemistry 1 SemesterDocumento24 paginePurification of Protiens: Mohan CC M.SC, Biochemistry 1 SemesterMelwin MejanNessuna valutazione finora
- Amino Acids and Proteins ReviewerDocumento11 pagineAmino Acids and Proteins ReviewerJohn-Karl JimenezNessuna valutazione finora
- Dna RnaDocumento42 pagineDna RnaSaumya DasNessuna valutazione finora
- Biochem Module 4 - Proteins and StructureDocumento15 pagineBiochem Module 4 - Proteins and StructureAnothando GobaNessuna valutazione finora
- Silica Based Stationary PhasesDocumento25 pagineSilica Based Stationary PhasesPriyanka GuptaNessuna valutazione finora
- Indira Gandhi National Tribal University Amarkantak, M.P. 484886Documento41 pagineIndira Gandhi National Tribal University Amarkantak, M.P. 484886Akanksha LangiwarNessuna valutazione finora
- Biochemistry - Proteins-StructuresDocumento34 pagineBiochemistry - Proteins-StructuresserficasoNessuna valutazione finora
- Biochemistry LN05-1Documento14 pagineBiochemistry LN05-1Rahaf Al-muhtasebNessuna valutazione finora
- Principal Investigator Dr.S.K.KhareDocumento11 paginePrincipal Investigator Dr.S.K.KhareAmit BansalNessuna valutazione finora
- Lecture 3-ProteinsDocumento9 pagineLecture 3-ProteinsOminousCroakNessuna valutazione finora
- Proline and HydroxyprolineDocumento7 pagineProline and HydroxyprolineYutman Wa dagoNessuna valutazione finora
- Complex at IonDocumento31 pagineComplex at IonShamsuzzaman TanimNessuna valutazione finora
- Week 2Documento25 pagineWeek 2Nurullah MertelNessuna valutazione finora
- 2017 USNCO Local Exam: 1 SolutionsDocumento16 pagine2017 USNCO Local Exam: 1 SolutionsAnuki TodriaNessuna valutazione finora
- Physical Science: Quarter 1 - Module: Title: Biological MacromoleculesDocumento26 paginePhysical Science: Quarter 1 - Module: Title: Biological MacromoleculesTeacher MelNessuna valutazione finora
- Biomolecules GRADE 12Documento26 pagineBiomolecules GRADE 12Jericho Anthiel de GuzmanNessuna valutazione finora
- BIOMOLECULESDocumento26 pagineBIOMOLECULESVicky VigneshNessuna valutazione finora
- Macromolecules Cut and Paste 2Documento5 pagineMacromolecules Cut and Paste 2Crazy BasketballNessuna valutazione finora
- Dimensionality Reduction For Protein Secondary Structure PredictionDocumento68 pagineDimensionality Reduction For Protein Secondary Structure PredictionAhmet EmreNessuna valutazione finora
- VRsec BIOINFORMATICSDocumento2 pagineVRsec BIOINFORMATICSKudipudi SrinivasNessuna valutazione finora
- Biology Syllabus For Integrated M.SC Course - Niser Semester 1Documento11 pagineBiology Syllabus For Integrated M.SC Course - Niser Semester 1Samyabrata SahaNessuna valutazione finora
- Recent Advances in The Synthesis and Applications of Collagen BasedDocumento45 pagineRecent Advances in The Synthesis and Applications of Collagen BasedJesús Alejandro Claudio RizoNessuna valutazione finora
- 11a Revision Pack T1Documento36 pagine11a Revision Pack T1Najat BteichNessuna valutazione finora
- MACROmoleculesDocumento80 pagineMACROmoleculesMaKenJi EscalanteNessuna valutazione finora
- EssentialsBiophysics PDFDocumento1 paginaEssentialsBiophysics PDFspd bahril100% (1)
- Lesson 4: The Chemistry of Carbon, The Chemistry of LifeDocumento6 pagineLesson 4: The Chemistry of Carbon, The Chemistry of LifeJGHUNGERNessuna valutazione finora
- In Silico Sequence Analysis, Homology Modeling and Function Annotation of Ocimum Basilicum Hypothetical Protein G1CT28 - OCIBADocumento8 pagineIn Silico Sequence Analysis, Homology Modeling and Function Annotation of Ocimum Basilicum Hypothetical Protein G1CT28 - OCIBAmariohuangNessuna valutazione finora
- Exam 1 AnswersDocumento9 pagineExam 1 AnswersA'Khris Fell-For Your-TypeNessuna valutazione finora
- Final Problems ACM - ICPC Hanoi 2012Documento14 pagineFinal Problems ACM - ICPC Hanoi 2012Phạm Duy HòaNessuna valutazione finora
- BSC Biochemistry Syllabus CBCSDocumento20 pagineBSC Biochemistry Syllabus CBCSRama Krishna SaiNessuna valutazione finora
- 9 BiomoleculesDocumento24 pagine9 BiomoleculesCBSE123.CO.NR100% (3)
- BSC Biotechnology: Choice Based Credit System See: Semester End ExaminationDocumento13 pagineBSC Biotechnology: Choice Based Credit System See: Semester End ExaminationNISHANTHNessuna valutazione finora
- A Majorcan Algebrist in King RNA's CourtDocumento41 pagineA Majorcan Algebrist in King RNA's CourtCesc RosselloNessuna valutazione finora
- Protein Structure and FunctionDocumento30 pagineProtein Structure and Functionمحمد جانNessuna valutazione finora
- Biology HL Study MaterialDocumento90 pagineBiology HL Study MaterialelenaNessuna valutazione finora
- Aqa A-Level Biology Cheatsheet PDFDocumento17 pagineAqa A-Level Biology Cheatsheet PDFLiza Khan100% (6)
- Protein SequencingDocumento5 pagineProtein SequencingSurajit BhattacharjeeNessuna valutazione finora
- Protein PredictionDocumento100 pagineProtein Predictionvani_darlingNessuna valutazione finora
- RISHUDocumento30 pagineRISHURishu Mittal100% (1)
- Introduction & BiomoleculesDocumento50 pagineIntroduction & BiomoleculesAshrul NasirNessuna valutazione finora
- WheyDocumento91 pagineWheyEviTrianaNessuna valutazione finora
- 1960 Brewer R. y Sleeman J.R. Soil Structure and Fabric Their Definition and DescriptionDocumento14 pagine1960 Brewer R. y Sleeman J.R. Soil Structure and Fabric Their Definition and DescriptionRonald LandaNessuna valutazione finora
- DNA Presentation StructuralDocumento25 pagineDNA Presentation StructuralAme Grace DubeNessuna valutazione finora
- The Nature of Drugs Vol. 1: History, Pharmacology, and Social ImpactDa EverandThe Nature of Drugs Vol. 1: History, Pharmacology, and Social ImpactValutazione: 5 su 5 stelle5/5 (5)
- Periodic Tales: A Cultural History of the Elements, from Arsenic to ZincDa EverandPeriodic Tales: A Cultural History of the Elements, from Arsenic to ZincValutazione: 3.5 su 5 stelle3.5/5 (137)
- AP® Chemistry Crash Course, For the 2020 Exam, Book + Online: Get a Higher Score in Less TimeDa EverandAP® Chemistry Crash Course, For the 2020 Exam, Book + Online: Get a Higher Score in Less TimeValutazione: 5 su 5 stelle5/5 (1)
- It's Elemental: The Hidden Chemistry in EverythingDa EverandIt's Elemental: The Hidden Chemistry in EverythingValutazione: 4 su 5 stelle4/5 (10)
- Is That a Fact?: Frauds, Quacks, and the Real Science of Everyday LifeDa EverandIs That a Fact?: Frauds, Quacks, and the Real Science of Everyday LifeValutazione: 5 su 5 stelle5/5 (4)
- Taste: Surprising Stories and Science About Why Food Tastes GoodDa EverandTaste: Surprising Stories and Science About Why Food Tastes GoodValutazione: 3 su 5 stelle3/5 (20)
- Guidelines for Defining Process Safety Competency RequirementsDa EverandGuidelines for Defining Process Safety Competency RequirementsValutazione: 3 su 5 stelle3/5 (1)
- Monkeys, Myths, and Molecules: Separating Fact from Fiction, and the Science of Everyday LifeDa EverandMonkeys, Myths, and Molecules: Separating Fact from Fiction, and the Science of Everyday LifeValutazione: 4 su 5 stelle4/5 (1)
- Chemistry for Breakfast: The Amazing Science of Everyday LifeDa EverandChemistry for Breakfast: The Amazing Science of Everyday LifeValutazione: 4.5 su 5 stelle4.5/5 (14)
- Formulating, Packaging, and Marketing of Natural Cosmetic ProductsDa EverandFormulating, Packaging, and Marketing of Natural Cosmetic ProductsNessuna valutazione finora
- Handbook of Formulating Dermal Applications: A Definitive Practical GuideDa EverandHandbook of Formulating Dermal Applications: A Definitive Practical GuideNessuna valutazione finora
- The Periodic Table: A Very Short IntroductionDa EverandThe Periodic Table: A Very Short IntroductionValutazione: 4.5 su 5 stelle4.5/5 (3)
- AP Chemistry Flashcards, Fourth Edition: Up-to-Date Review and PracticeDa EverandAP Chemistry Flashcards, Fourth Edition: Up-to-Date Review and PracticeNessuna valutazione finora
- The Billion-Dollar Molecule: The Quest for the Perfect DrugDa EverandThe Billion-Dollar Molecule: The Quest for the Perfect DrugValutazione: 5 su 5 stelle5/5 (2)
- Guidelines for Integrating Process Safety into Engineering ProjectsDa EverandGuidelines for Integrating Process Safety into Engineering ProjectsNessuna valutazione finora
- Fundamentals of Chemistry: A Modern IntroductionDa EverandFundamentals of Chemistry: A Modern IntroductionValutazione: 5 su 5 stelle5/5 (1)
- Chemistry for Breakfast: The Amazing Science of Everyday LifeDa EverandChemistry for Breakfast: The Amazing Science of Everyday LifeValutazione: 4.5 su 5 stelle4.5/5 (90)
- The Nature of Drugs Vol. 1: History, Pharmacology, and Social ImpactDa EverandThe Nature of Drugs Vol. 1: History, Pharmacology, and Social ImpactValutazione: 5 su 5 stelle5/5 (1)
- Organic Chemistry for Schools: Advanced Level and Senior High SchoolDa EverandOrganic Chemistry for Schools: Advanced Level and Senior High SchoolNessuna valutazione finora
- Tribology: Friction and Wear of Engineering MaterialsDa EverandTribology: Friction and Wear of Engineering MaterialsValutazione: 5 su 5 stelle5/5 (1)
- The Disappearing Spoon: And Other True Tales of Madness, Love, and the History of the World from the Periodic Table of the ElementsDa EverandThe Disappearing Spoon: And Other True Tales of Madness, Love, and the History of the World from the Periodic Table of the ElementsValutazione: 4 su 5 stelle4/5 (146)